Riddhish Bhalodia
The Llama 4 Herd: Architecture, Training, Evaluation, and Deployment Notes
Jan 15, 2026Abstract:This document consolidates publicly reported technical details about Metas Llama 4 model family. It summarizes (i) released variants (Scout and Maverick) and the broader herd context including the previewed Behemoth teacher model, (ii) architectural characteristics beyond a high-level MoE description covering routed/shared-expert structure, early-fusion multimodality, and long-context design elements reported for Scout (iRoPE and length generalization strategies), (iii) training disclosures spanning pre-training, mid-training for long-context extension, and post-training methodology (lightweight SFT, online RL, and lightweight DPO) as described in release materials, (iv) developer-reported benchmark results for both base and instruction-tuned checkpoints, and (v) practical deployment constraints observed across major serving environments, including provider-specific context limits and quantization packaging. The manuscript also summarizes licensing obligations relevant to redistribution and derivative naming, and reviews publicly described safeguards and evaluation practices. The goal is to provide a compact technical reference for researchers and practitioners who need precise, source-backed facts about Llama 4.
GenCA: A Text-conditioned Generative Model for Realistic and Drivable Codec Avatars
Aug 24, 2024

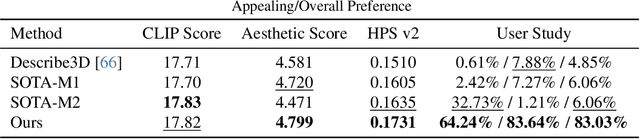
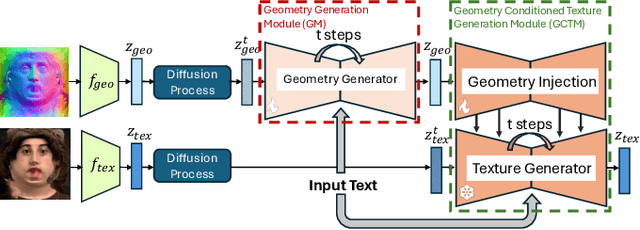
Abstract:Photo-realistic and controllable 3D avatars are crucial for various applications such as virtual and mixed reality (VR/MR), telepresence, gaming, and film production. Traditional methods for avatar creation often involve time-consuming scanning and reconstruction processes for each avatar, which limits their scalability. Furthermore, these methods do not offer the flexibility to sample new identities or modify existing ones. On the other hand, by learning a strong prior from data, generative models provide a promising alternative to traditional reconstruction methods, easing the time constraints for both data capture and processing. Additionally, generative methods enable downstream applications beyond reconstruction, such as editing and stylization. Nonetheless, the research on generative 3D avatars is still in its infancy, and therefore current methods still have limitations such as creating static avatars, lacking photo-realism, having incomplete facial details, or having limited drivability. To address this, we propose a text-conditioned generative model that can generate photo-realistic facial avatars of diverse identities, with more complete details like hair, eyes and mouth interior, and which can be driven through a powerful non-parametric latent expression space. Specifically, we integrate the generative and editing capabilities of latent diffusion models with a strong prior model for avatar expression driving. Our model can generate and control high-fidelity avatars, even those out-of-distribution. We also highlight its potential for downstream applications, including avatar editing and single-shot avatar reconstruction.
RENs: Relevance Encoding Networks
May 25, 2022



Abstract:The manifold assumption for high-dimensional data assumes that the data is generated by varying a set of parameters obtained from a low-dimensional latent space. Deep generative models (DGMs) are widely used to learn data representations in an unsupervised way. DGMs parameterize the underlying low-dimensional manifold in the data space using bottleneck architectures such as variational autoencoders (VAEs). The bottleneck dimension for VAEs is treated as a hyperparameter that depends on the dataset and is fixed at design time after extensive tuning. As the intrinsic dimensionality of most real-world datasets is unknown, often, there is a mismatch between the intrinsic dimensionality and the latent dimensionality chosen as a hyperparameter. This mismatch can negatively contribute to the model performance for representation learning and sample generation tasks. This paper proposes relevance encoding networks (RENs): a novel probabilistic VAE-based framework that uses the automatic relevance determination (ARD) prior in the latent space to learn the data-specific bottleneck dimensionality. The relevance of each latent dimension is directly learned from the data along with the other model parameters using stochastic gradient descent and a reparameterization trick adapted to non-Gaussian priors. We leverage the concept of DeepSets to capture permutation invariant statistical properties in both data and latent spaces for relevance determination. The proposed framework is general and flexible and can be used for the state-of-the-art VAE models that leverage regularizers to impose specific characteristics in the latent space (e.g., disentanglement). With extensive experimentation on synthetic and public image datasets, we show that the proposed model learns the relevant latent bottleneck dimensionality without compromising the representation and generation quality of the samples.
Learning Population-level Shape Statistics and Anatomy Segmentation From Images: A Joint Deep Learning Model
Jan 10, 2022



Abstract:Statistical shape modeling is an essential tool for the quantitative analysis of anatomical populations. Point distribution models (PDMs) represent the anatomical surface via a dense set of correspondences, an intuitive and easy-to-use shape representation for subsequent applications. These correspondences are exhibited in two coordinate spaces: the local coordinates describing the geometrical features of each individual anatomical surface and the world coordinates representing the population-level statistical shape information after removing global alignment differences across samples in the given cohort. We propose a deep-learning-based framework that simultaneously learns these two coordinate spaces directly from the volumetric images. The proposed joint model serves a dual purpose; the world correspondences can directly be used for shape analysis applications, circumventing the heavy pre-processing and segmentation involved in traditional PDM models. Additionally, the local correspondences can be used for anatomy segmentation. We demonstrate the efficacy of this joint model for both shape modeling applications on two datasets and its utility in inferring the anatomical surface.
Leveraging Unsupervised Image Registration for Discovery of Landmark Shape Descriptor
Nov 13, 2021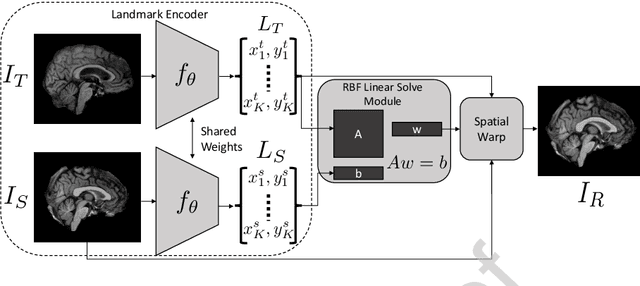
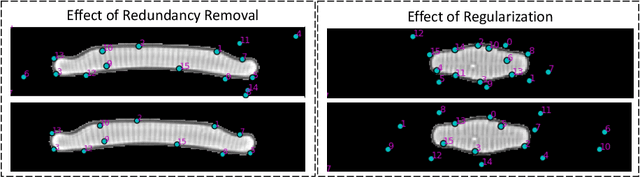
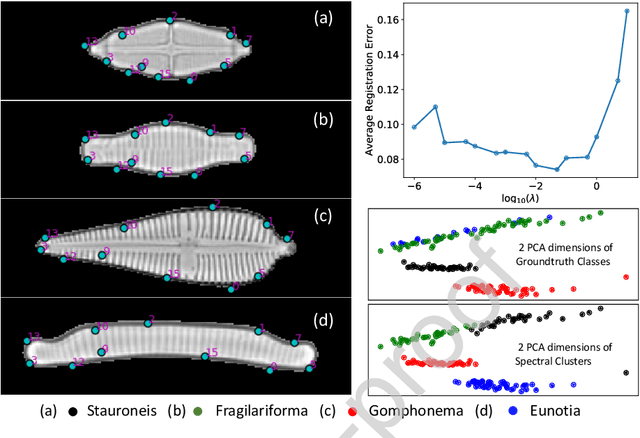
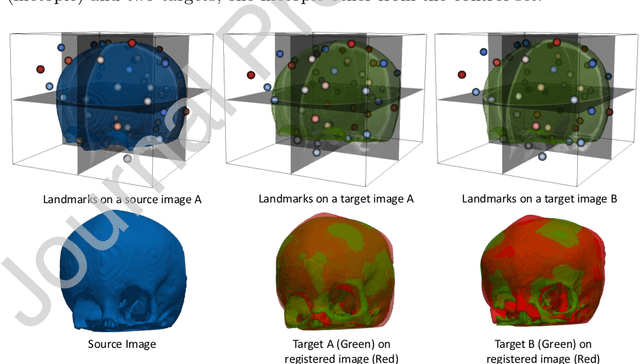
Abstract:In current biological and medical research, statistical shape modeling (SSM) provides an essential framework for the characterization of anatomy/morphology. Such analysis is often driven by the identification of a relatively small number of geometrically consistent features found across the samples of a population. These features can subsequently provide information about the population shape variation. Dense correspondence models can provide ease of computation and yield an interpretable low-dimensional shape descriptor when followed by dimensionality reduction. However, automatic methods for obtaining such correspondences usually require image segmentation followed by significant preprocessing, which is taxing in terms of both computation as well as human resources. In many cases, the segmentation and subsequent processing require manual guidance and anatomy specific domain expertise. This paper proposes a self-supervised deep learning approach for discovering landmarks from images that can directly be used as a shape descriptor for subsequent analysis. We use landmark-driven image registration as the primary task to force the neural network to discover landmarks that register the images well. We also propose a regularization term that allows for robust optimization of the neural network and ensures that the landmarks uniformly span the image domain. The proposed method circumvents segmentation and preprocessing and directly produces a usable shape descriptor using just 2D or 3D images. In addition, we also propose two variants on the training loss function that allows for prior shape information to be integrated into the model. We apply this framework on several 2D and 3D datasets to obtain their shape descriptors, and analyze their utility for various applications.
DeepSSM: A Blueprint for Image-to-Shape Deep Learning Models
Oct 14, 2021

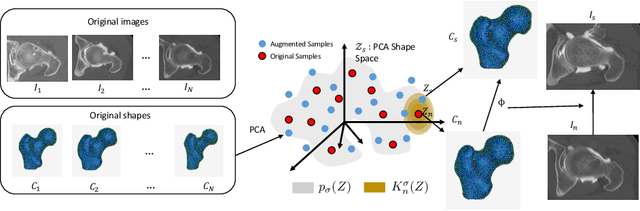
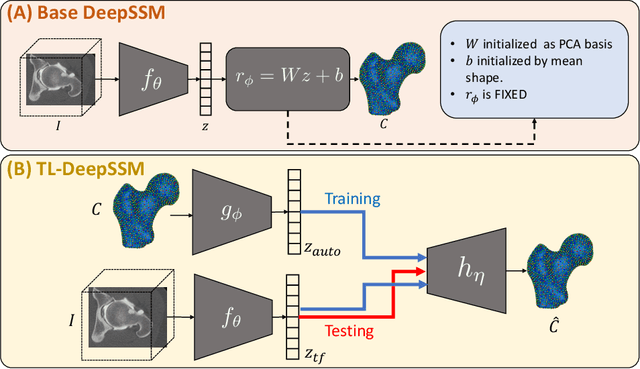
Abstract:Statistical shape modeling (SSM) characterizes anatomical variations in a population of shapes generated from medical images. SSM requires consistent shape representation across samples in shape cohort. Establishing this representation entails a processing pipeline that includes anatomy segmentation, re-sampling, registration, and non-linear optimization. These shape representations are then used to extract low-dimensional shape descriptors that facilitate subsequent analyses in different applications. However, the current process of obtaining these shape descriptors from imaging data relies on human and computational resources, requiring domain expertise for segmenting anatomies of interest. Moreover, this same taxing pipeline needs to be repeated to infer shape descriptors for new image data using a pre-trained/existing shape model. Here, we propose DeepSSM, a deep learning-based framework for learning the functional mapping from images to low-dimensional shape descriptors and their associated shape representations, thereby inferring statistical representation of anatomy directly from 3D images. Once trained using an existing shape model, DeepSSM circumvents the heavy and manual pre-processing and segmentation and significantly improves the computational time, making it a viable solution for fully end-to-end SSM applications. In addition, we introduce a model-based data-augmentation strategy to address data scarcity. Finally, this paper presents and analyzes two different architectural variants of DeepSSM with different loss functions using three medical datasets and their downstream clinical application. Experiments showcase that DeepSSM performs comparably or better to the state-of-the-art SSM both quantitatively and on application-driven downstream tasks. Therefore, DeepSSM aims to provide a comprehensive blueprint for deep learning-based image-to-shape models.
Improving Pneumonia Localization via Cross-Attention on Medical Images and Reports
Oct 06, 2021



Abstract:Localization and characterization of diseases like pneumonia are primary steps in a clinical pipeline, facilitating detailed clinical diagnosis and subsequent treatment planning. Additionally, such location annotated datasets can provide a pathway for deep learning models to be used for downstream tasks. However, acquiring quality annotations is expensive on human resources and usually requires domain expertise. On the other hand, medical reports contain a plethora of information both about pneumonia characteristics and its location. In this paper, we propose a novel weakly-supervised attention-driven deep learning model that leverages encoded information in medical reports during training to facilitate better localization. Our model also performs classification of attributes that are associated to pneumonia and extracted from medical reports for supervision. Both the classification and localization are trained in conjunction and once trained, the model can be utilized for both the localization and characterization of pneumonia using only the input image. In this paper, we explore and analyze the model using chest X-ray datasets and demonstrate qualitatively and quantitatively that the introduction of textual information improves pneumonia localization. We showcase quantitative results on two datasets, MIMIC-CXR and Chest X-ray-8, and we also showcase severity characterization on the COVID-19 dataset.
Unsupervised Shape Normality Metric for Severity Quantification
Jul 18, 2020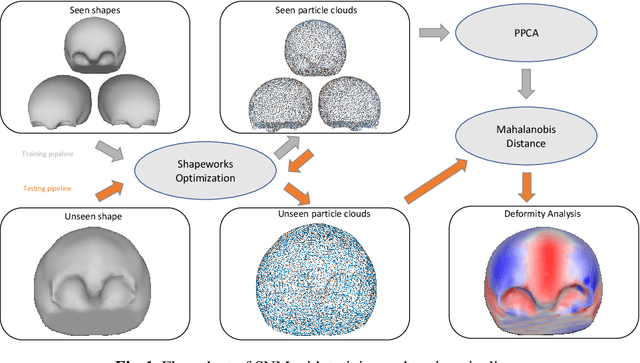
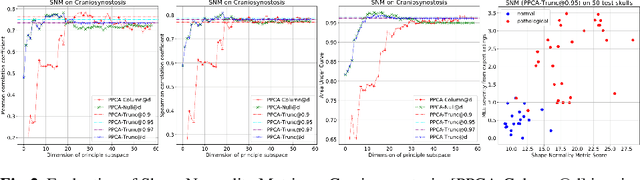

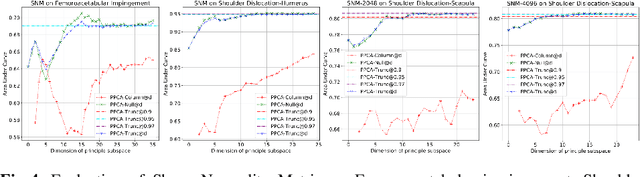
Abstract:This work describes an unsupervised method to objectively quantify the abnormality of general anatomical shapes. The severity of an anatomical deformity often serves as a determinant in the clinical management of patients. However, experiential bias and distinctive random residuals among specialist individuals bring variability in diagnosis and patient management decisions, irrespective of the objective deformity degree. Therefore, supervised methods are prone to be misled given insufficient labeling of pathological samples that inevitably preserve human bias and inconsistency. Furthermore, subjects demonstrating a specific pathology are naturally rare relative to the normal population. To avoid relying on sufficient pathological samples by fully utilizing the power of normal samples, we propose the shape normality metric (SNM), which requires learning only from normal samples and zero knowledge about the pathology. We represent shapes by landmarks automatically inferred from the data and model the normal group by a multivariate Gaussian distribution. Extensive experiments on different anatomical datasets, including skulls, femurs, scapulae, and humeri, demonstrate that SNM can provide an effective normality measurement, which can significantly detect and indicate pathology. Therefore, SNM offers promising value in a variety of clinical applications.
Uncertain-DeepSSM: From Images to Probabilistic Shape Models
Jul 13, 2020



Abstract:Statistical shape modeling (SSM) has recently taken advantage of advances in deep learning to alleviate the need for a time-consuming and expert-driven workflow of anatomy segmentation, shape registration, and the optimization of population-level shape representations. DeepSSM is an end-to-end deep learning approach that extracts statistical shape representation directly from unsegmented images with little manual overhead. It performs comparably with state-of-the-art shape modeling methods for estimating morphologies that are viable for subsequent downstream tasks. Nonetheless, DeepSSM produces an overconfident estimate of shape that cannot be blindly assumed to be accurate. Hence, conveying what DeepSSM does not know, via quantifying granular estimates of uncertainty, is critical for its direct clinical application as an on-demand diagnostic tool to determine how trustworthy the model output is. Here, we propose Uncertain-DeepSSM as a unified model that quantifies both, data-dependent aleatoric uncertainty by adapting the network to predict intrinsic input variance, and model-dependent epistemic uncertainty via a Monte Carlo dropout sampling to approximate a variational distribution over the network parameters. Experiments show an accuracy improvement over DeepSSM while maintaining the same benefits of being end-to-end with little pre-processing.
Self-Supervised Discovery of Anatomical Shape Landmarks
Jun 13, 2020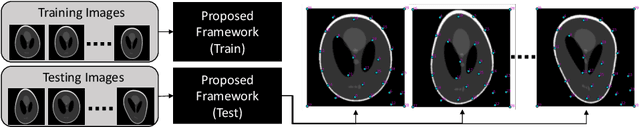
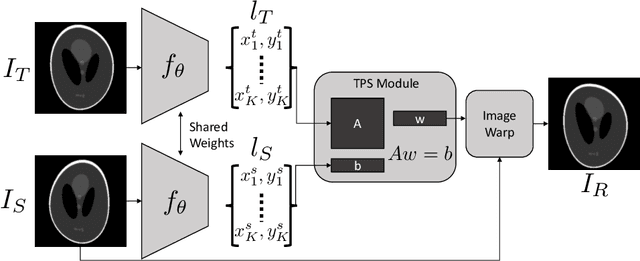


Abstract:Statistical shape analysis is a very useful tool in a wide range of medical and biological applications. However, it typically relies on the ability to produce a relatively small number of features that can capture the relevant variability in a population. State-of-the-art methods for obtaining such anatomical features rely on either extensive preprocessing or segmentation and/or significant tuning and post-processing. These shortcomings limit the widespread use of shape statistics. We propose that effective shape representations should provide sufficient information to align/register images. Using this assumption we propose a self-supervised, neural network approach for automatically positioning and detecting landmarks in images that can be used for subsequent analysis. The network discovers the landmarks corresponding to anatomical shape features that promote good image registration in the context of a particular class of transformations. In addition, we also propose a regularization for the proposed network which allows for a uniform distribution of these discovered landmarks. In this paper, we present a complete framework, which only takes a set of input images and produces landmarks that are immediately usable for statistical shape analysis. We evaluate the performance on a phantom dataset as well as 2D and 3D images.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge