Irene Vignon-Clementel
SIMBIOTX
Multi-fidelity graph-based neural networks architectures to learn Navier-Stokes solutions on non-parametrized 2D domains
Jan 05, 2026Abstract:We propose a graph-based, multi-fidelity learning framework for the prediction of stationary Navier--Stokes solutions in non-parametrized two-dimensional geometries. The method is designed to guide the learning process through successive approximations, starting from reduced-order and full Stokes models, and progressively approaching the Navier--Stokes solution. To effectively capture both local and long-range dependencies in the velocity and pressure fields, we combine graph neural networks with Transformer and Mamba architectures. While Transformers achieve the highest accuracy, we show that Mamba can be successfully adapted to graph-structured data through an unsupervised node-ordering strategy. The Mamba approach significantly reduces computational cost while maintaining performance. Physical knowledge is embedded directly into the architecture through an encoding -- processing -- physics informed decoding pipeline. Derivatives are computed through algebraic operators constructed via the Weighted Least Squares method. The flexibility of these operators allows us not only to make the output obey the governing equations, but also to constrain selected hidden features to satisfy mass conservation. We introduce additional physical biases through an enriched graph convolution with the same differential operators describing the PDEs. Overall, we successfully guide the learning process by physical knowledge and fluid dynamics insights, leading to more regular and accurate predictions
Enhancing the automatic segmentation and analysis of 3D liver vasculature models
Nov 24, 2024Abstract:Surgical assessment of liver cancer patients requires identification of the vessel trees from medical images. Specifically, the venous trees - the portal (perfusing) and the hepatic (draining) trees are important for understanding the liver anatomy and disease state, and perform surgery planning. This research aims to improve the 3D segmentation, skeletonization, and subsequent analysis of vessel trees, by creating an automatic pipeline based on deep learning and image processing techniques. The first part of this work explores the impact of differentiable skeletonization methods such as ClDice and morphological skeletonization loss, on the overall liver vessel segmentation performance. To this aim, it studies how to improve vessel tree connectivity. The second part of this study converts a single class vessel segmentation into multi-class ones, separating the two venous trees. It builds on the previous two-class vessel segmentation model, which vessel tree outputs might be entangled, and on connected components and skeleton analyses of the trees. After providing sub-labeling of the specific anatomical branches of each venous tree, these algorithms also enable a morphometric analysis of the vessel trees by extracting various geometrical markers. In conclusion, we propose a method that successfully improves current skeletonization methods, for extensive vascular trees that contain vessels of different calibers. The separation algorithm creates a clean multi-class segmentation of the vessels, validated by surgeons to provide low error. A new, publicly shared high-quality liver vessel dataset of 77 cases is thus created. Finally a method to annotate vessel trees according to anatomy is provided, enabling a unique liver vessel morphometry analysis.
CoRe: An Automated Pipeline for The Prediction of Liver Resection Complexity from Preoperative CT Scans
Oct 15, 2022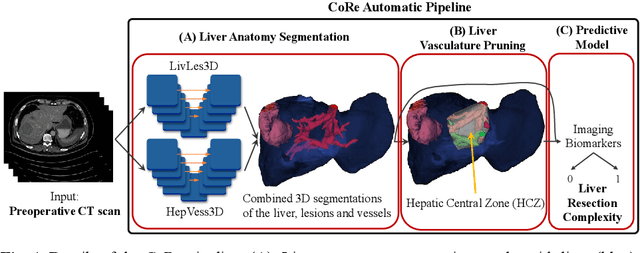
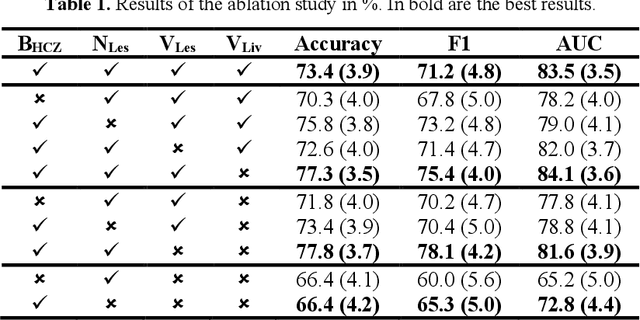
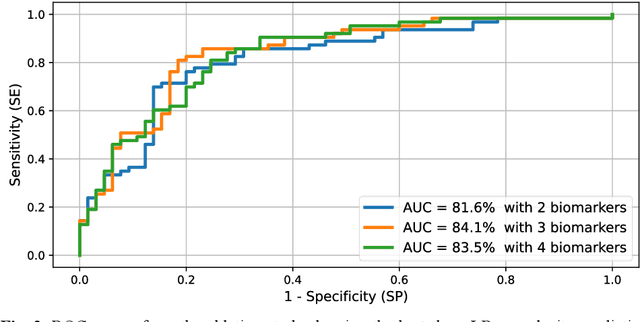

Abstract:Surgical resections are the most prevalent curative treatment for primary liver cancer. Tumors located in critical positions are known to complexify liver resections (LR). While experienced surgeons in specialized medical centers may have the necessary expertise to accurately anticipate LR complexity, and prepare accordingly, an objective method able to reproduce this behavior would have the potential to improve the standard routine of care, and avoid intra- and postoperative complications. In this article, we propose CoRe, an automated medical image processing pipeline for the prediction of postoperative LR complexity from preoperative CT scans, using imaging biomarkers. The CoRe pipeline first segments the liver, lesions, and vessels with two deep learning networks. The liver vasculature is then pruned based on a topological criterion to define the hepatic central zone (HCZ), a convex volume circumscribing the major liver vessels, from which a new imaging biomarker, BHCZ is derived. Additional biomarkers are extracted and leveraged to train and evaluate a LR complexity prediction model. An ablation study shows the HCZ-based biomarker as the central feature in predicting LR complexity. The best predictive model reaches an accuracy, F1, and AUC of 77.3, 75.4, and 84.1% respectively.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge