Geeticka Chauhan
Training Large ASR Encoders with Differential Privacy
Sep 21, 2024



Abstract:Self-supervised learning (SSL) methods for large speech models have proven to be highly effective at ASR. With the interest in public deployment of large pre-trained models, there is a rising concern for unintended memorization and leakage of sensitive data points from the training data. In this paper, we apply differentially private (DP) pre-training to a SOTA Conformer-based encoder, and study its performance on a downstream ASR task assuming the fine-tuning data is public. This paper is the first to apply DP to SSL for ASR, investigating the DP noise tolerance of the BEST-RQ pre-training method. Notably, we introduce a novel variant of model pruning called gradient-based layer freezing that provides strong improvements in privacy-utility-compute trade-offs. Our approach yields a LibriSpeech test-clean/other WER (%) of 3.78/ 8.41 with ($10$, 1e^-9)-DP for extrapolation towards low dataset scales, and 2.81/ 5.89 with (10, 7.9e^-11)-DP for extrapolation towards high scales.
Bidirectional Captioning for Clinically Accurate and Interpretable Models
Oct 30, 2023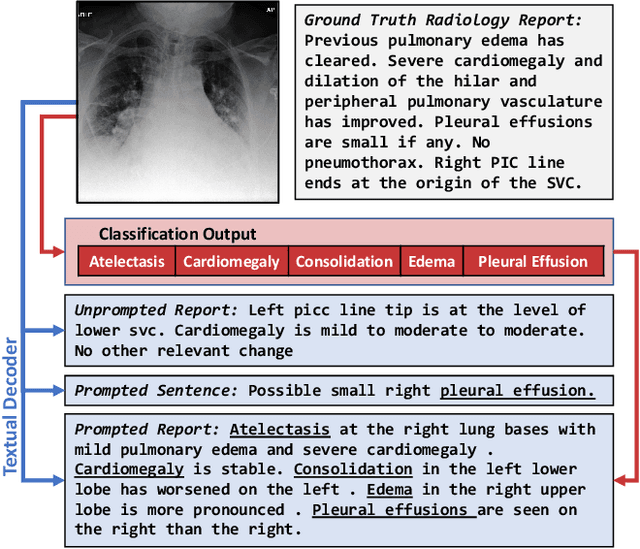
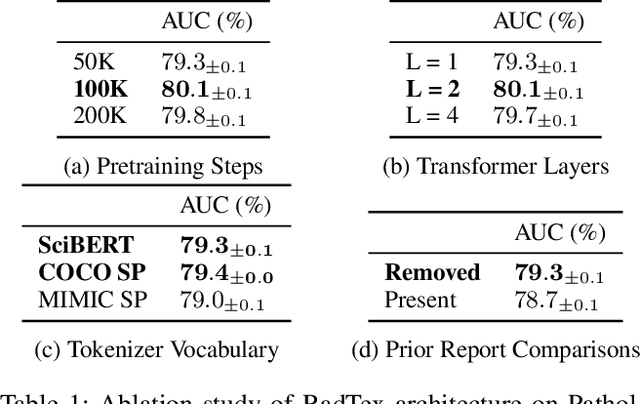
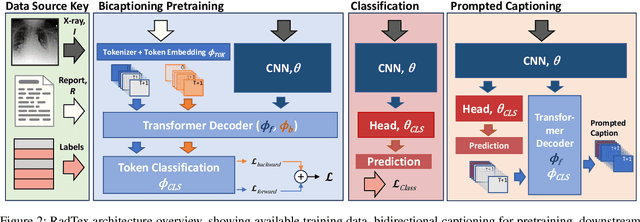
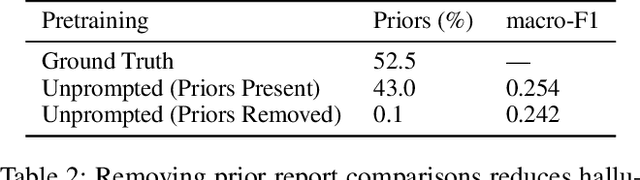
Abstract:Vision-language pretraining has been shown to produce high-quality visual encoders which transfer efficiently to downstream computer vision tasks. While generative language models have gained widespread attention, image captioning has thus far been mostly overlooked as a form of cross-modal pretraining in favor of contrastive learning, especially in medical image analysis. In this paper, we experiment with bidirectional captioning of radiology reports as a form of pretraining and compare the quality and utility of learned embeddings with those from contrastive pretraining methods. We optimize a CNN encoder, transformer decoder architecture named RadTex for the radiology domain. Results show that not only does captioning pretraining yield visual encoders that are competitive with contrastive pretraining (CheXpert competition multi-label AUC of 89.4%), but also that our transformer decoder is capable of generating clinically relevant reports (captioning macro-F1 score of 0.349 using CheXpert labeler) and responding to prompts with targeted, interactive outputs.
RadTex: Learning Efficient Radiograph Representations from Text Reports
Aug 05, 2022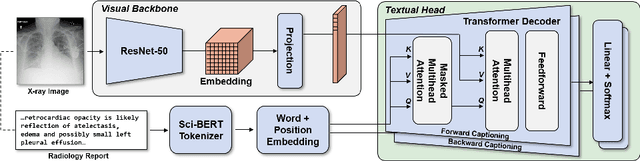
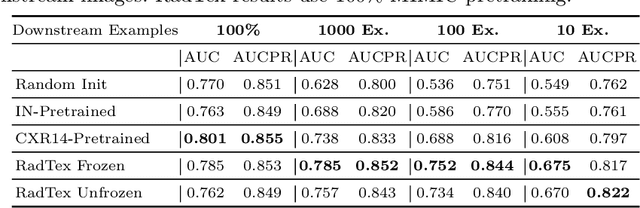
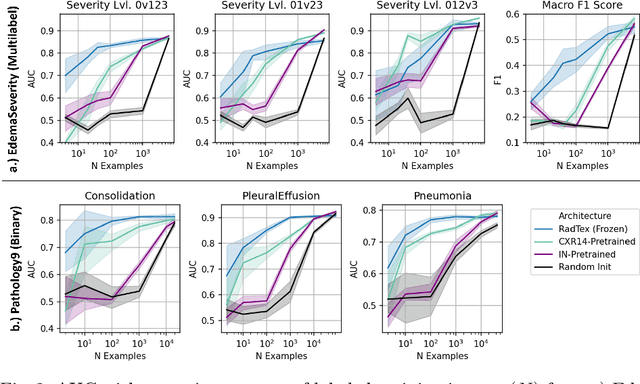
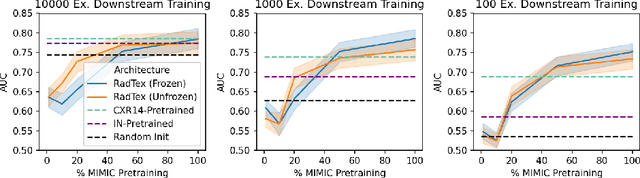
Abstract:Automated analysis of chest radiography using deep learning has tremendous potential to enhance the clinical diagnosis of diseases in patients. However, deep learning models typically require large amounts of annotated data to achieve high performance -- often an obstacle to medical domain adaptation. In this paper, we build a data-efficient learning framework that utilizes radiology reports to improve medical image classification performance with limited labeled data (fewer than 1000 examples). Specifically, we examine image-captioning pretraining to learn high-quality medical image representations that train on fewer examples. Following joint pretraining of a convolutional encoder and transformer decoder, we transfer the learned encoder to various classification tasks. Averaged over 9 pathologies, we find that our model achieves higher classification performance than ImageNet-supervised and in-domain supervised pretraining when labeled training data is limited.
Explainable Deep Learning in Healthcare: A Methodological Survey from an Attribution View
Dec 05, 2021
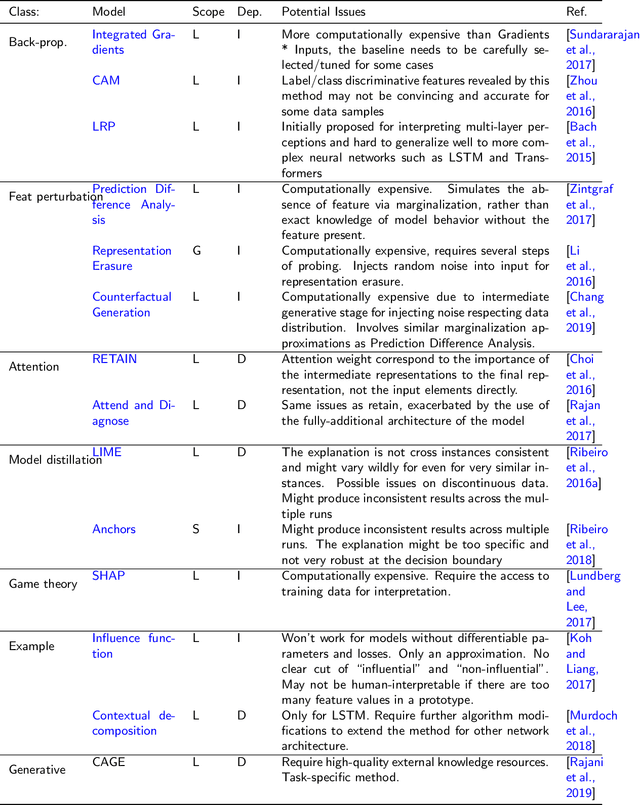
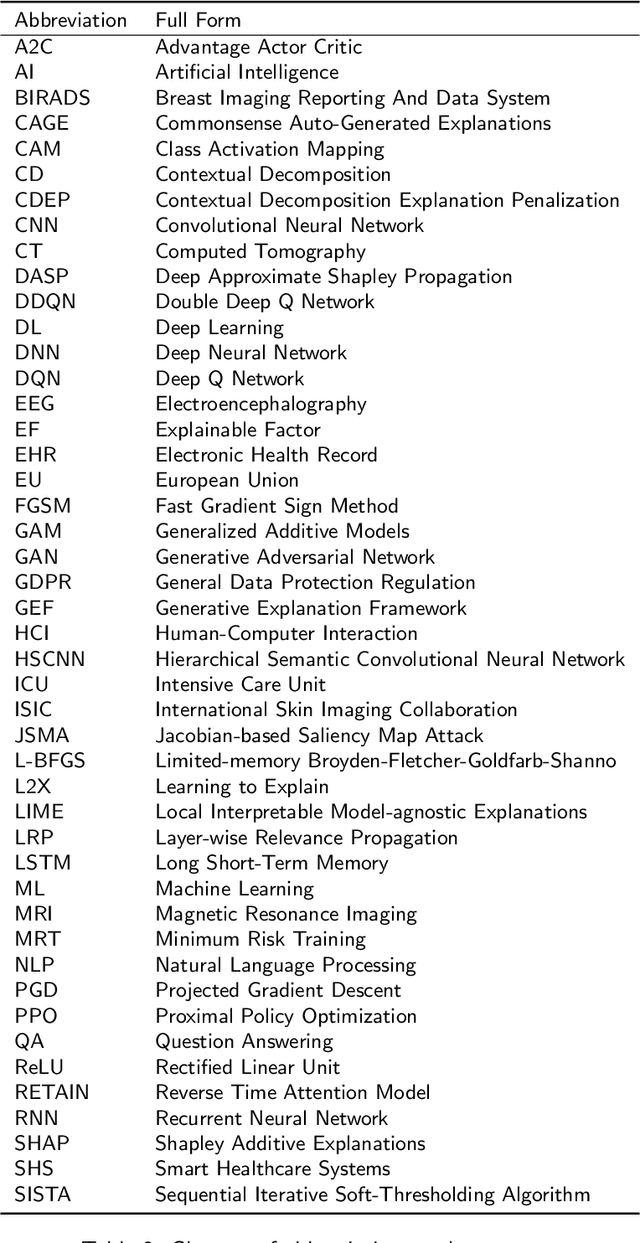
Abstract:The increasing availability of large collections of electronic health record (EHR) data and unprecedented technical advances in deep learning (DL) have sparked a surge of research interest in developing DL based clinical decision support systems for diagnosis, prognosis, and treatment. Despite the recognition of the value of deep learning in healthcare, impediments to further adoption in real healthcare settings remain due to the black-box nature of DL. Therefore, there is an emerging need for interpretable DL, which allows end users to evaluate the model decision making to know whether to accept or reject predictions and recommendations before an action is taken. In this review, we focus on the interpretability of the DL models in healthcare. We start by introducing the methods for interpretability in depth and comprehensively as a methodological reference for future researchers or clinical practitioners in this field. Besides the methods' details, we also include a discussion of advantages and disadvantages of these methods and which scenarios each of them is suitable for, so that interested readers can know how to compare and choose among them for use. Moreover, we discuss how these methods, originally developed for solving general-domain problems, have been adapted and applied to healthcare problems and how they can help physicians better understand these data-driven technologies. Overall, we hope this survey can help researchers and practitioners in both artificial intelligence (AI) and clinical fields understand what methods we have for enhancing the interpretability of their DL models and choose the optimal one accordingly.
How Good Is NLP? A Sober Look at NLP Tasks through the Lens of Social Impact
Jun 04, 2021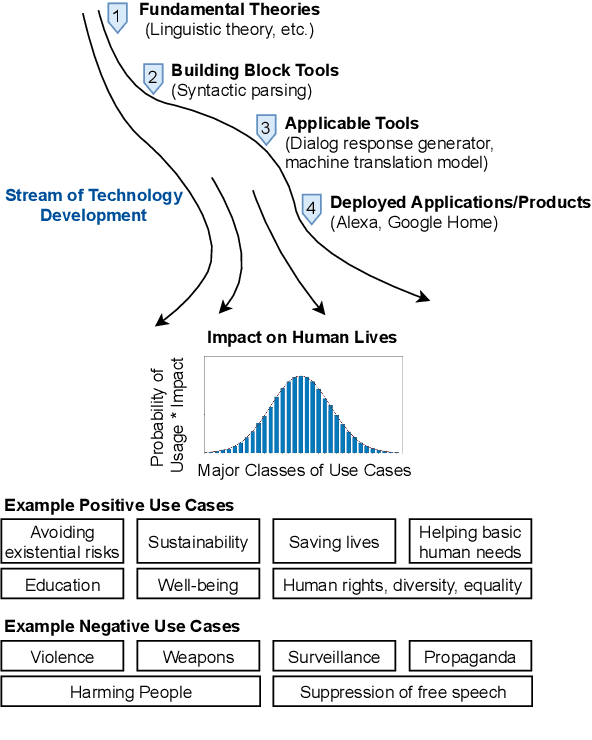
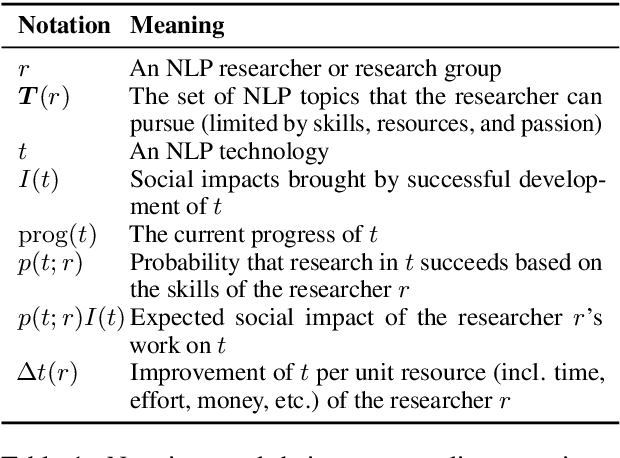
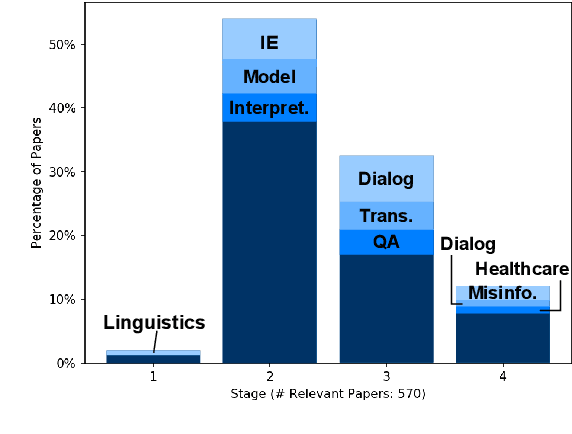
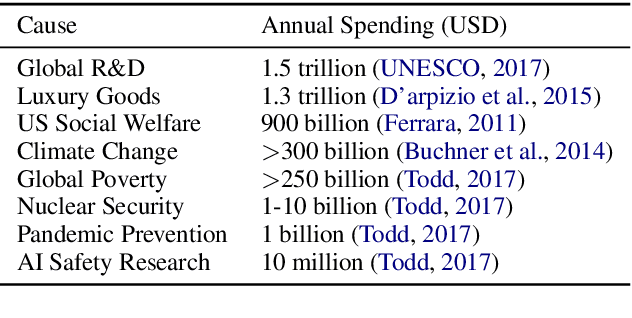
Abstract:Recent years have seen many breakthroughs in natural language processing (NLP), transitioning it from a mostly theoretical field to one with many real-world applications. Noting the rising number of applications of other machine learning and AI techniques with pervasive societal impact, we anticipate the rising importance of developing NLP technologies for social good. Inspired by theories in moral philosophy and global priorities research, we aim to promote a guideline for social good in the context of NLP. We lay the foundations via moral philosophy's definition of social good, propose a framework to evaluate NLP tasks' direct and indirect real-world impact, and adopt the methodology of global priorities research to identify priority causes for NLP research. Finally, we use our theoretical framework to provide some practical guidelines for future NLP research for social good. Our data and codes are available at http://github.com/zhijing-jin/nlp4sg_acl2021
Joint Modeling of Chest Radiographs and Radiology Reports for Pulmonary Edema Assessment
Aug 22, 2020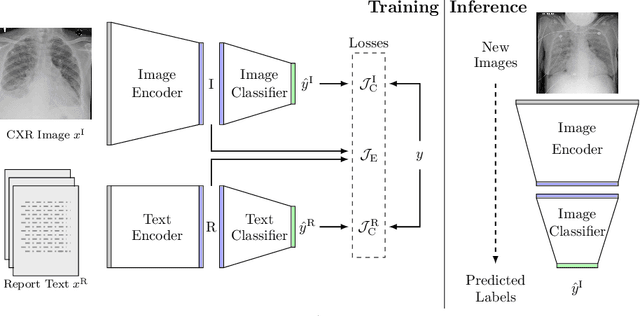
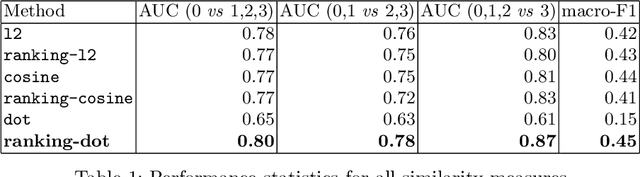

Abstract:We propose and demonstrate a novel machine learning algorithm that assesses pulmonary edema severity from chest radiographs. While large publicly available datasets of chest radiographs and free-text radiology reports exist, only limited numerical edema severity labels can be extracted from radiology reports. This is a significant challenge in learning such models for image classification. To take advantage of the rich information present in the radiology reports, we develop a neural network model that is trained on both images and free-text to assess pulmonary edema severity from chest radiographs at inference time. Our experimental results suggest that the joint image-text representation learning improves the performance of pulmonary edema assessment compared to a supervised model trained on images only. We also show the use of the text for explaining the image classification by the joint model. To the best of our knowledge, our approach is the first to leverage free-text radiology reports for improving the image model performance in this application. Our code is available at https://github.com/RayRuizhiLiao/joint_chestxray.
REflex: Flexible Framework for Relation Extraction in Multiple Domains
Jul 20, 2019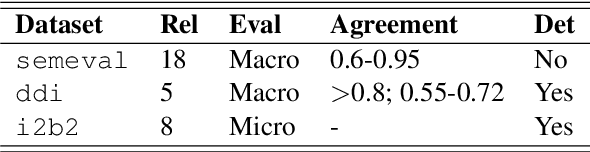
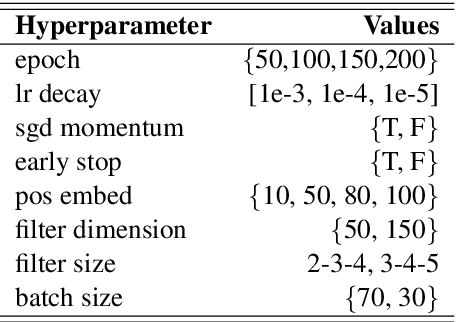
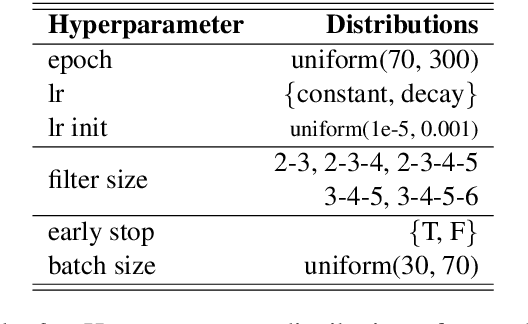
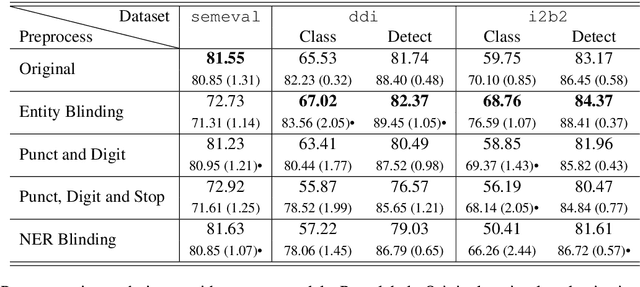
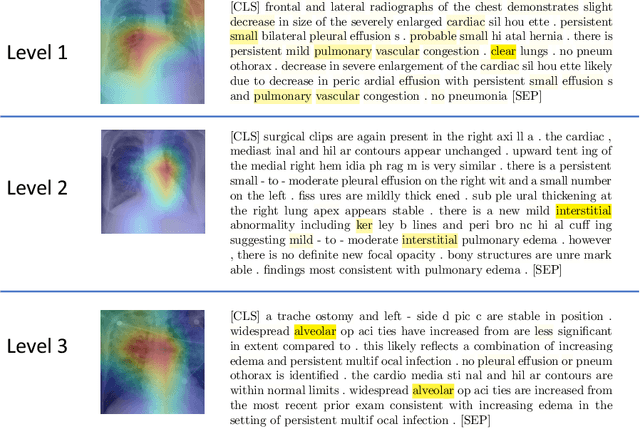
Abstract:Systematic comparison of methods for relation extraction (RE) is difficult because many experiments in the field are not described precisely enough to be completely reproducible and many papers fail to report ablation studies that would highlight the relative contributions of their various combined techniques. In this work, we build a unifying framework for RE, applying this on three highly used datasets (from the general, biomedical and clinical domains) with the ability to be extendable to new datasets. By performing a systematic exploration of modeling, pre-processing and training methodologies, we find that choices of pre-processing are a large contributor performance and that omission of such information can further hinder fair comparison. Other insights from our exploration allow us to provide recommendations for future research in this area.
MIMIC-Extract: A Data Extraction, Preprocessing, and Representation Pipeline for MIMIC-III
Jul 19, 2019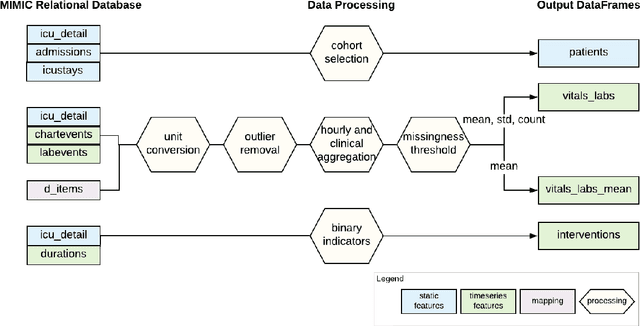
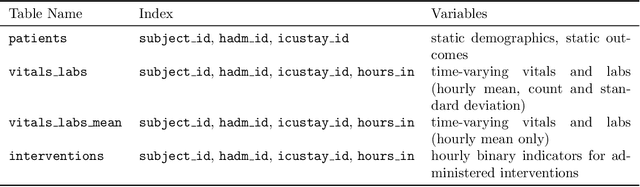
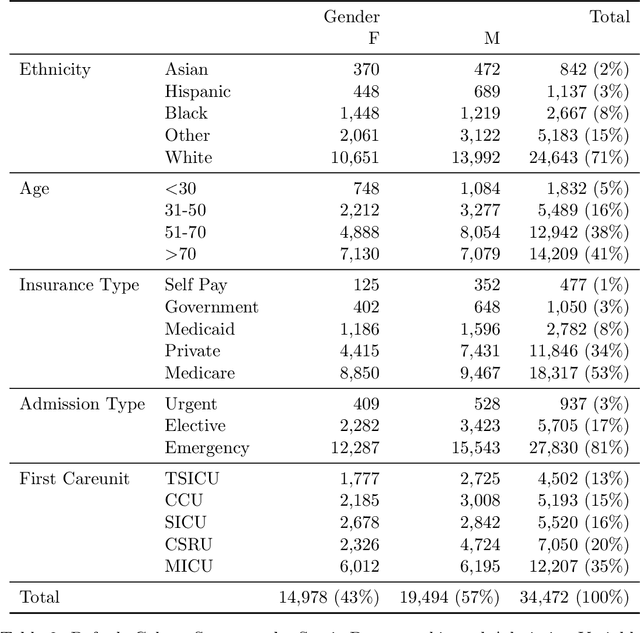
Abstract:Robust machine learning relies on access to data that can be used with standardized frameworks in important tasks and the ability to develop models whose performance can be reasonably reproduced. In machine learning for healthcare, the community faces reproducibility challenges due to a lack of publicly accessible data and a lack of standardized data processing frameworks. We present MIMIC-Extract, an open-source pipeline for transforming raw electronic health record (EHR) data for critical care patients contained in the publicly-available MIMIC-III database into dataframes that are directly usable in common machine learning pipelines. MIMIC-Extract addresses three primary challenges in making complex health records data accessible to the broader machine learning community. First, it provides standardized data processing functions, including unit conversion, outlier detection, and aggregating semantically equivalent features, thus accounting for duplication and reducing missingness. Second, it preserves the time series nature of clinical data and can be easily integrated into clinically actionable prediction tasks in machine learning for health. Finally, it is highly extensible so that other researchers with related questions can easily use the same pipeline. We demonstrate the utility of this pipeline by showcasing several benchmark tasks and baseline results.
Rethinking clinical prediction: Why machine learning must consider year of care and feature aggregation
Nov 30, 2018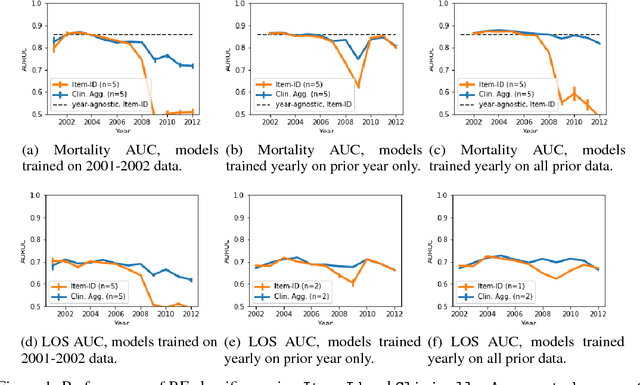
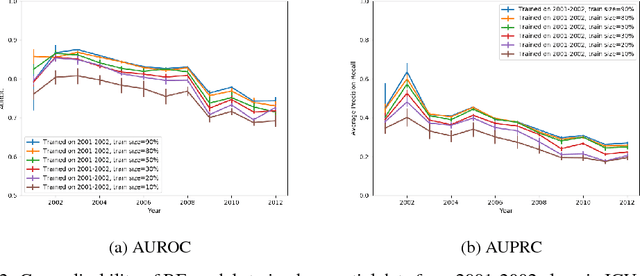
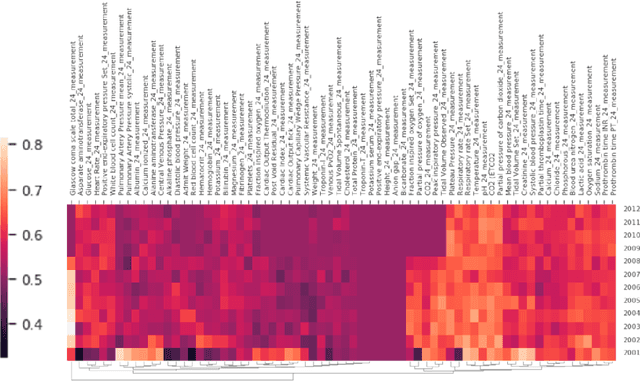
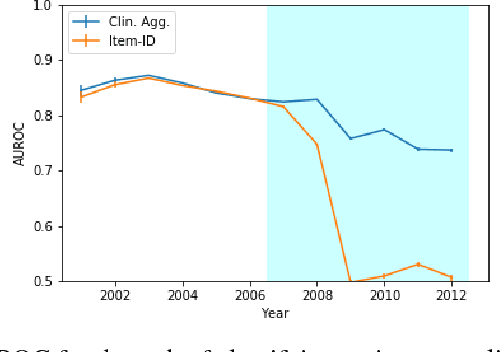
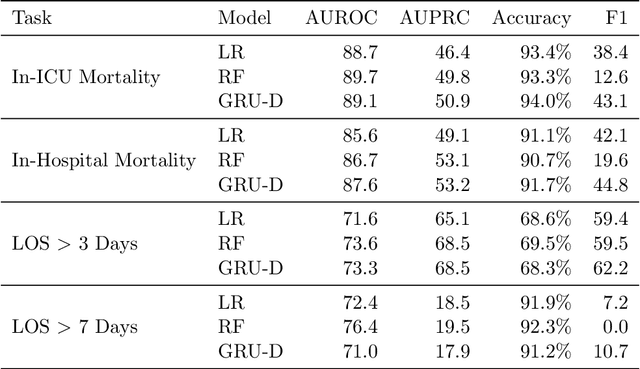
Abstract:Machine learning for healthcare often trains models on de-identified datasets with randomly-shifted calendar dates, ignoring the fact that data were generated under hospital operation practices that change over time. These changing practices induce definitive changes in observed data which confound evaluations which do not account for dates and limit the generalisability of date-agnostic models. In this work, we establish the magnitude of this problem on MIMIC, a public hospital dataset, and showcase a simple solution. We augment MIMIC with the year in which care was provided and show that a model trained using standard feature representations will significantly degrade in quality over time. We find a deterioration of 0.3 AUC when evaluating mortality prediction on data from 10 years later. We find a similar deterioration of 0.15 AUC for length-of-stay. In contrast, we demonstrate that clinically-oriented aggregates of raw features significantly mitigate future deterioration. Our suggested aggregated representations, when retrained yearly, have prediction quality comparable to year-agnostic models.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge