Anna Goldenberg
University of Toronto, Vector Institute, SickKids
DynaSubVAE: Adaptive Subgrouping for Scalable and Robust OOD Detection
Jun 11, 2025Abstract:Real-world observational data often contain existing or emerging heterogeneous subpopulations that deviate from global patterns. The majority of models tend to overlook these underrepresented groups, leading to inaccurate or even harmful predictions. Existing solutions often rely on detecting these samples as Out-of-domain (OOD) rather than adapting the model to new emerging patterns. We introduce DynaSubVAE, a Dynamic Subgrouping Variational Autoencoder framework that jointly performs representation learning and adaptive OOD detection. Unlike conventional approaches, DynaSubVAE evolves with the data by dynamically updating its latent structure to capture new trends. It leverages a novel non-parametric clustering mechanism, inspired by Gaussian Mixture Models, to discover and model latent subgroups based on embedding similarity. Extensive experiments show that DynaSubVAE achieves competitive performance in both near-OOD and far-OOD detection, and excels in class-OOD scenarios where an entire class is missing during training. We further illustrate that our dynamic subgrouping mechanism outperforms standalone clustering methods such as GMM and KMeans++ in terms of both OOD accuracy and regret precision.
ExOSITO: Explainable Off-Policy Learning with Side Information for Intensive Care Unit Blood Test Orders
Apr 24, 2025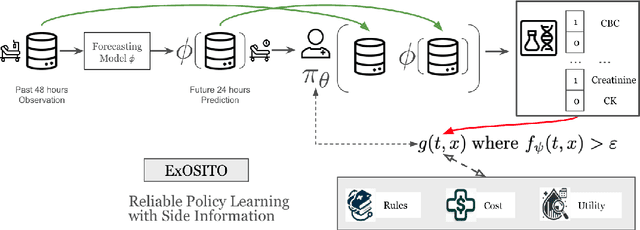
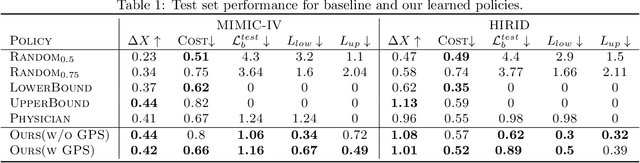
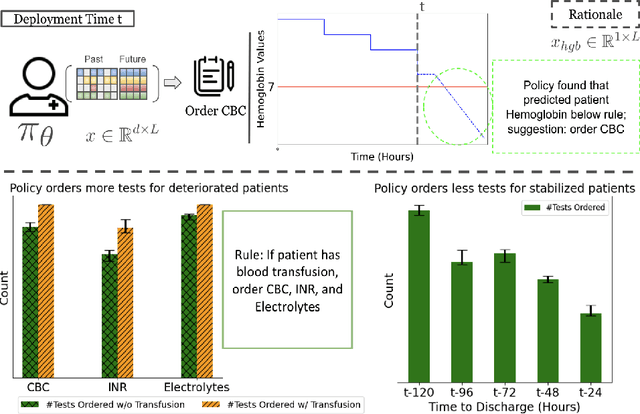

Abstract:Ordering a minimal subset of lab tests for patients in the intensive care unit (ICU) can be challenging. Care teams must balance between ensuring the availability of the right information and reducing the clinical burden and costs associated with each lab test order. Most in-patient settings experience frequent over-ordering of lab tests, but are now aiming to reduce this burden on both hospital resources and the environment. This paper develops a novel method that combines off-policy learning with privileged information to identify the optimal set of ICU lab tests to order. Our approach, EXplainable Off-policy learning with Side Information for ICU blood Test Orders (ExOSITO) creates an interpretable assistive tool for clinicians to order lab tests by considering both the observed and predicted future status of each patient. We pose this problem as a causal bandit trained using offline data and a reward function derived from clinically-approved rules; we introduce a novel learning framework that integrates clinical knowledge with observational data to bridge the gap between the optimal and logging policies. The learned policy function provides interpretable clinical information and reduces costs without omitting any vital lab orders, outperforming both a physician's policy and prior approaches to this practical problem.
Measurement Scheduling for ICU Patients with Offline Reinforcement Learning
Feb 12, 2024Abstract:Scheduling laboratory tests for ICU patients presents a significant challenge. Studies show that 20-40% of lab tests ordered in the ICU are redundant and could be eliminated without compromising patient safety. Prior work has leveraged offline reinforcement learning (Offline-RL) to find optimal policies for ordering lab tests based on patient information. However, new ICU patient datasets have since been released, and various advancements have been made in Offline-RL methods. In this study, we first introduce a preprocessing pipeline for the newly-released MIMIC-IV dataset geared toward time-series tasks. We then explore the efficacy of state-of-the-art Offline-RL methods in identifying better policies for ICU patient lab test scheduling. Besides assessing methodological performance, we also discuss the overall suitability and practicality of using Offline-RL frameworks for scheduling laboratory tests in ICU settings.
Learning from Time Series under Temporal Label Noise
Feb 06, 2024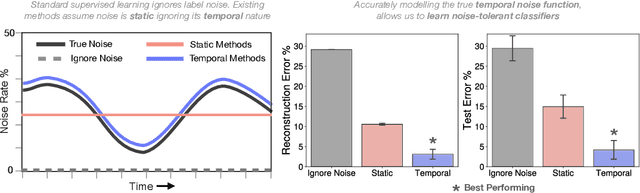
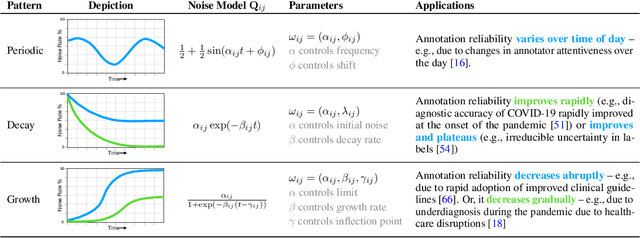
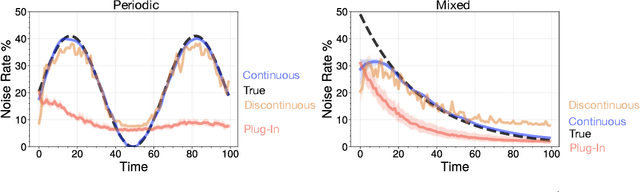
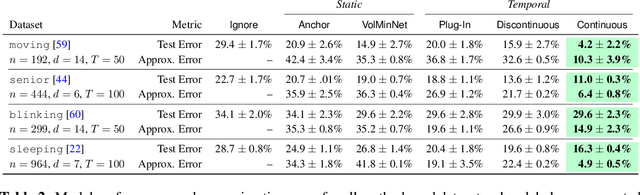
Abstract:Many sequential classification tasks are affected by label noise that varies over time. Such noise can cause label quality to improve, worsen, or periodically change over time. We first propose and formalize temporal label noise, an unstudied problem for sequential classification of time series. In this setting, multiple labels are recorded in sequence while being corrupted by a time-dependent noise function. We first demonstrate the importance of modelling the temporal nature of the label noise function and how existing methods will consistently underperform. We then propose methods that can train noise-tolerant classifiers by estimating the temporal label noise function directly from data. We show that our methods lead to state-of-the-art performance in the presence of diverse temporal label noise functions using real and synthetic data.
Integrate Any Omics: Towards genome-wide data integration for patient stratification
Jan 15, 2024Abstract:High-throughput omics profiling advancements have greatly enhanced cancer patient stratification. However, incomplete data in multi-omics integration presents a significant challenge, as traditional methods like sample exclusion or imputation often compromise biological diversity and dependencies. Furthermore, the critical task of accurately classifying new patients with partial omics data into existing subtypes is commonly overlooked. To address these issues, we introduce IntegrAO (Integrate Any Omics), an unsupervised framework for integrating incomplete multi-omics data and classifying new samples. IntegrAO first combines partially overlapping patient graphs from diverse omics sources and utilizes graph neural networks to produce unified patient embeddings. Our systematic evaluation across five cancer cohorts involving six omics modalities demonstrates IntegrAO's robustness to missing data and its accuracy in classifying new samples with partial profiles. An acute myeloid leukemia case study further validates its capability to uncover biological and clinical heterogeneity in incomplete datasets. IntegrAO's ability to handle heterogeneous and incomplete data makes it an essential tool for precision oncology, offering a holistic approach to patient characterization.
Maintaining Stability and Plasticity for Predictive Churn Reduction
May 06, 2023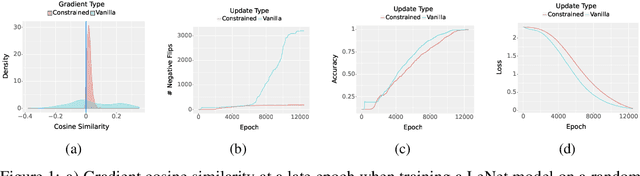
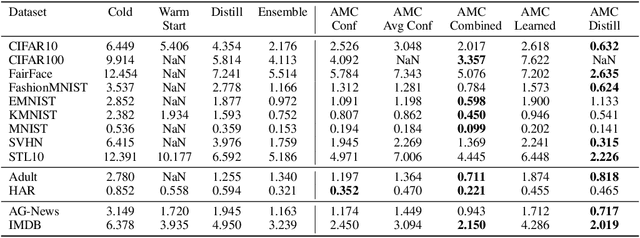
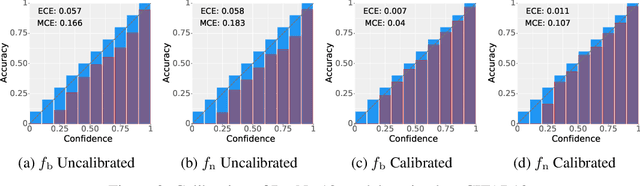
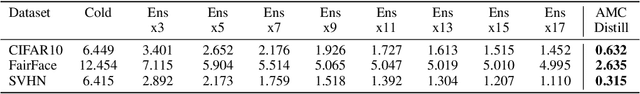
Abstract:Deployed machine learning models should be updated to take advantage of a larger sample size to improve performance, as more data is gathered over time. Unfortunately, even when model updates improve aggregate metrics such as accuracy, they can lead to errors on samples that were correctly predicted by the previous model causing per-sample regression in performance known as predictive churn. Such prediction flips erode user trust thereby reducing the effectiveness of the human-AI team as a whole. We propose a solution called Accumulated Model Combination (AMC) based keeping the previous and current model version, and generating a meta-output using the prediction of the two models. AMC is a general technique and we propose several instances of it, each having their own advantages depending on the model and data properties. AMC requires minimal additional computation and changes to training procedures. We motivate the need for AMC by showing the difficulty of making a single model consistent with its own predictions throughout training thereby revealing an implicit stability-plasticity tradeoff when training a single model. We demonstrate the effectiveness of AMC on a variety of modalities including computer vision, text, and tabular datasets comparing against state-of-the-art churn reduction methods, and showing superior churn reduction ability compared to all existing methods while being more efficient than ensembles.
From Single-Visit to Multi-Visit Image-Based Models: Single-Visit Models are Enough to Predict Obstructive Hydronephrosis
Dec 27, 2022Abstract:Previous work has shown the potential of deep learning to predict renal obstruction using kidney ultrasound images. However, these image-based classifiers have been trained with the goal of single-visit inference in mind. We compare methods from video action recognition (i.e. convolutional pooling, LSTM, TSM) to adapt single-visit convolutional models to handle multiple visit inference. We demonstrate that incorporating images from a patient's past hospital visits provides only a small benefit for the prediction of obstructive hydronephrosis. Therefore, inclusion of prior ultrasounds is beneficial, but prediction based on the latest ultrasound is sufficient for patient risk stratification.
Time-Varying Correlation Networks for Interpretable Change Point Detection
Nov 08, 2022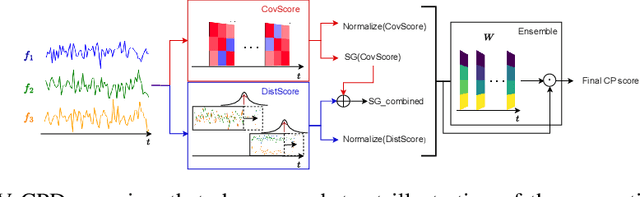
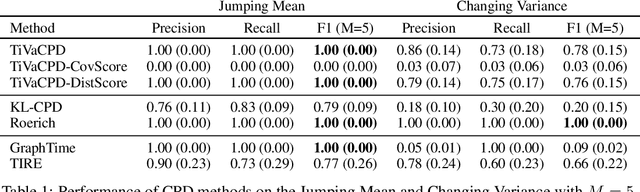
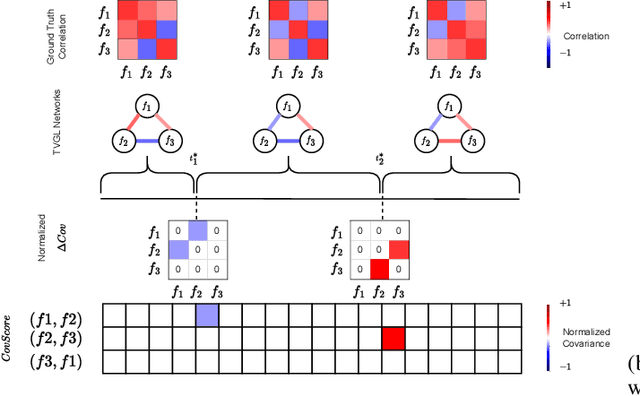
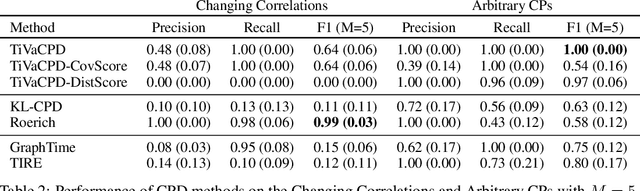
Abstract:Change point detection (CPD) methods aim to detect abrupt changes in time-series data. Recent CPD methods have demonstrated their potential in identifying changes in underlying statistical distributions but often fail to capture complex changes in the correlation structure in time-series data. These methods also fail to generalize effectively, as even within the same time-series, different kinds of change points (CPs) may arise that are best characterized by different types of time-series perturbations. To address this issue, we propose TiVaCPD, a CPD methodology that uses a time-varying graphical lasso based method to identify changes in correlation patterns between features over time, and combines that with an aggregate Kernel Maximum Mean Discrepancy (MMD) test to identify subtle changes in the underlying statistical distributions of dynamically established time windows. We evaluate the performance of TiVaCPD in identifying and characterizing various types of CPs in time-series and show that our method outperforms current state-of-the-art CPD methods for all categories of CPs.
Decoupling Local and Global Representations of Time Series
Feb 11, 2022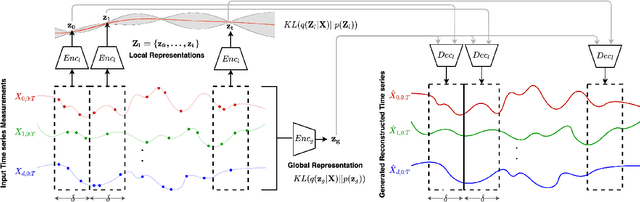
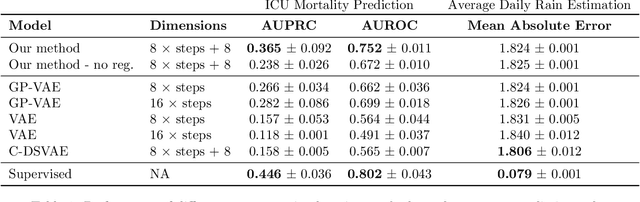
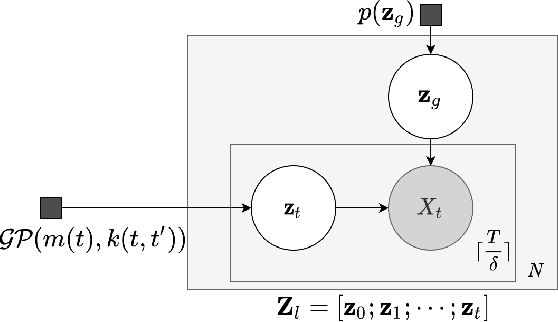
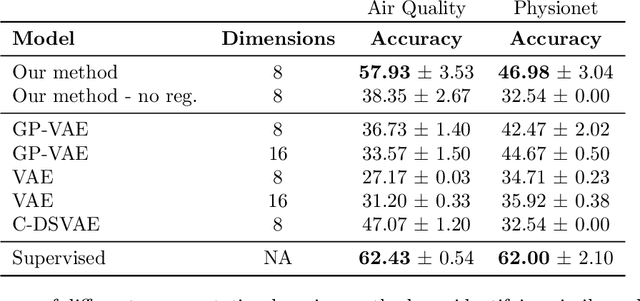
Abstract:Real-world time series data are often generated from several sources of variation. Learning representations that capture the factors contributing to this variability enables a better understanding of the data via its underlying generative process and improves performance on downstream machine learning tasks. This paper proposes a novel generative approach for learning representations for the global and local factors of variation in time series. The local representation of each sample models non-stationarity over time with a stochastic process prior, and the global representation of the sample encodes the time-independent characteristics. To encourage decoupling between the representations, we introduce counterfactual regularization that minimizes the mutual information between the two variables. In experiments, we demonstrate successful recovery of the true local and global variability factors on simulated data, and show that representations learned using our method yield superior performance on downstream tasks on real-world datasets. We believe that the proposed way of defining representations is beneficial for data modelling and yields better insights into the complexity of real-world data.
Extracting Clinician's Goals by What-if Interpretable Modeling
Oct 28, 2021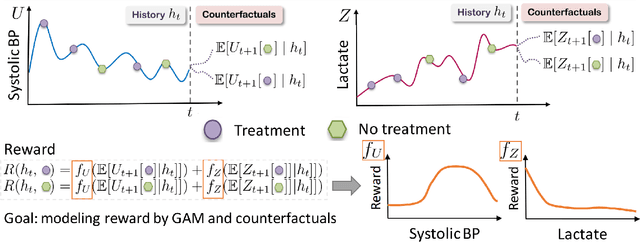
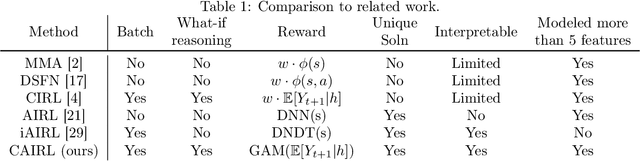
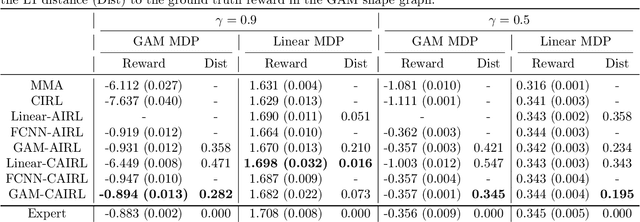
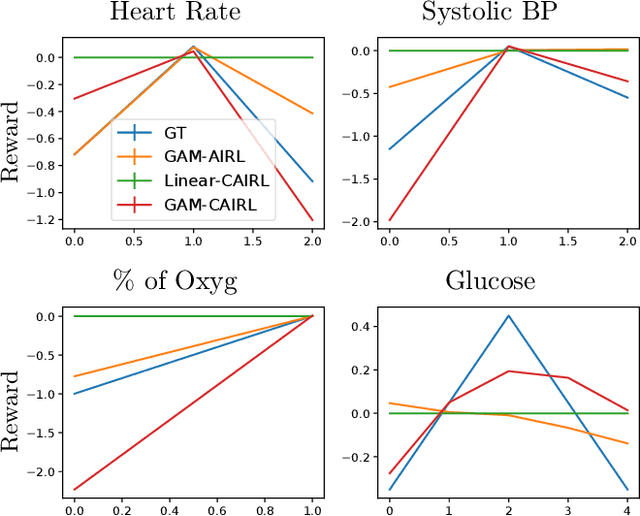
Abstract:Although reinforcement learning (RL) has tremendous success in many fields, applying RL to real-world settings such as healthcare is challenging when the reward is hard to specify and no exploration is allowed. In this work, we focus on recovering clinicians' rewards in treating patients. We incorporate the what-if reasoning to explain clinician's actions based on future outcomes. We use generalized additive models (GAMs) - a class of accurate, interpretable models - to recover the reward. In both simulation and a real-world hospital dataset, we show our model outperforms baselines. Finally, our model's explanations match several clinical guidelines when treating patients while we found the previously-used linear model often contradicts them.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge