Benjamin Haibe-Kains
Lassoed Forests: Random Forests with Adaptive Lasso Post-selection
Nov 11, 2025Abstract:Random forests are a statistical learning technique that use bootstrap aggregation to average high-variance and low-bias trees. Improvements to random forests, such as applying Lasso regression to the tree predictions, have been proposed in order to reduce model bias. However, these changes can sometimes degrade performance (e.g., an increase in mean squared error). In this paper, we show in theory that the relative performance of these two methods, standard and Lasso-weighted random forests, depends on the signal-to-noise ratio. We further propose a unified framework to combine random forests and Lasso selection by applying adaptive weighting and show mathematically that it can strictly outperform the other two methods. We compare the three methods through simulation, including bias-variance decomposition, error estimates evaluation, and variable importance analysis. We also show the versatility of our method by applications to a variety of real-world datasets.
Integrate Any Omics: Towards genome-wide data integration for patient stratification
Jan 15, 2024Abstract:High-throughput omics profiling advancements have greatly enhanced cancer patient stratification. However, incomplete data in multi-omics integration presents a significant challenge, as traditional methods like sample exclusion or imputation often compromise biological diversity and dependencies. Furthermore, the critical task of accurately classifying new patients with partial omics data into existing subtypes is commonly overlooked. To address these issues, we introduce IntegrAO (Integrate Any Omics), an unsupervised framework for integrating incomplete multi-omics data and classifying new samples. IntegrAO first combines partially overlapping patient graphs from diverse omics sources and utilizes graph neural networks to produce unified patient embeddings. Our systematic evaluation across five cancer cohorts involving six omics modalities demonstrates IntegrAO's robustness to missing data and its accuracy in classifying new samples with partial profiles. An acute myeloid leukemia case study further validates its capability to uncover biological and clinical heterogeneity in incomplete datasets. IntegrAO's ability to handle heterogeneous and incomplete data makes it an essential tool for precision oncology, offering a holistic approach to patient characterization.
Maintaining Stability and Plasticity for Predictive Churn Reduction
May 06, 2023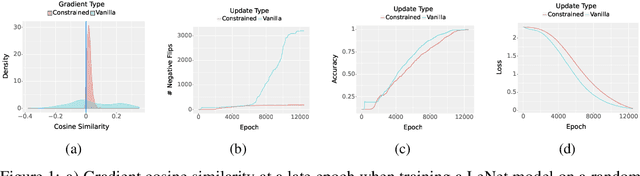
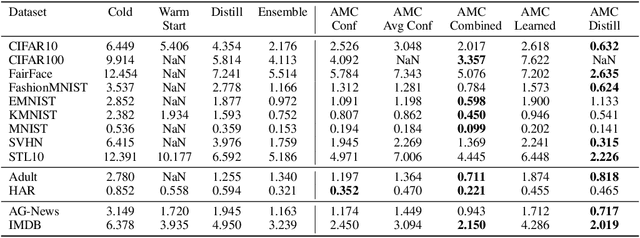
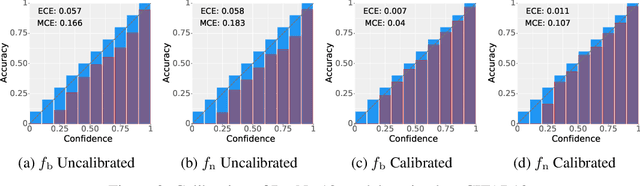
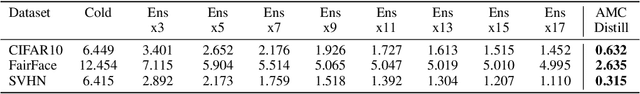
Abstract:Deployed machine learning models should be updated to take advantage of a larger sample size to improve performance, as more data is gathered over time. Unfortunately, even when model updates improve aggregate metrics such as accuracy, they can lead to errors on samples that were correctly predicted by the previous model causing per-sample regression in performance known as predictive churn. Such prediction flips erode user trust thereby reducing the effectiveness of the human-AI team as a whole. We propose a solution called Accumulated Model Combination (AMC) based keeping the previous and current model version, and generating a meta-output using the prediction of the two models. AMC is a general technique and we propose several instances of it, each having their own advantages depending on the model and data properties. AMC requires minimal additional computation and changes to training procedures. We motivate the need for AMC by showing the difficulty of making a single model consistent with its own predictions throughout training thereby revealing an implicit stability-plasticity tradeoff when training a single model. We demonstrate the effectiveness of AMC on a variety of modalities including computer vision, text, and tabular datasets comparing against state-of-the-art churn reduction methods, and showing superior churn reduction ability compared to all existing methods while being more efficient than ensembles.
FastCPH: Efficient Survival Analysis for Neural Networks
Aug 21, 2022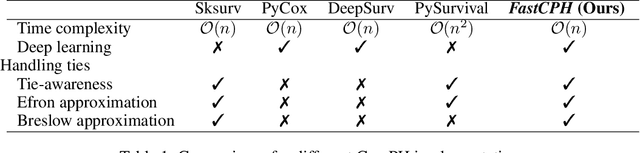
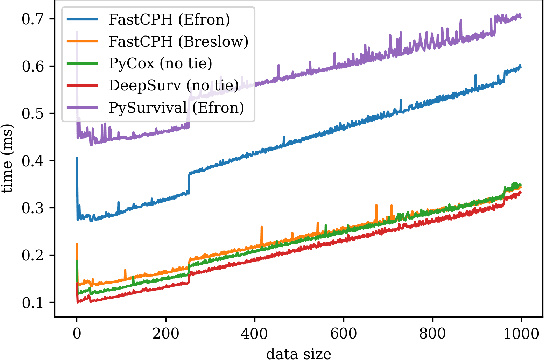
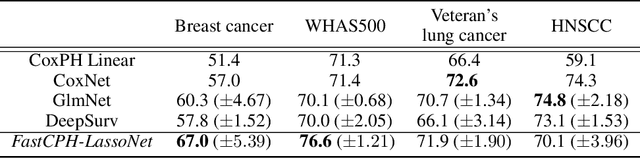
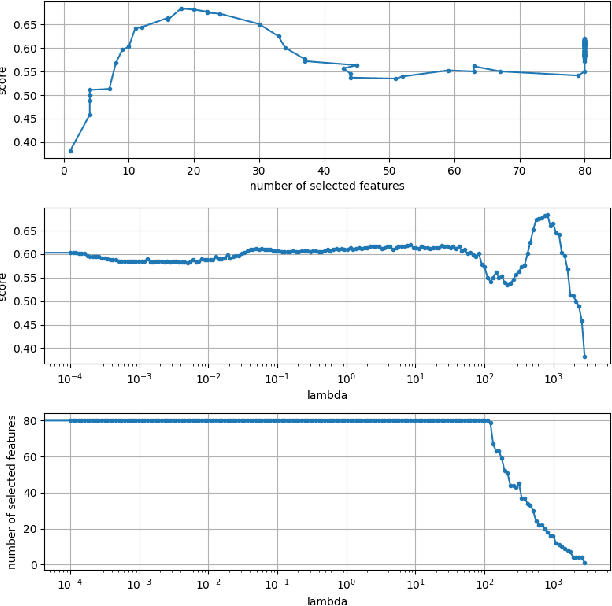
Abstract:The Cox proportional hazards model is a canonical method in survival analysis for prediction of the life expectancy of a patient given clinical or genetic covariates -- it is a linear model in its original form. In recent years, several methods have been proposed to generalize the Cox model to neural networks, but none of these are both numerically correct and computationally efficient. We propose FastCPH, a new method that runs in linear time and supports both the standard Breslow and Efron methods for tied events. We also demonstrate the performance of FastCPH combined with LassoNet, a neural network that provides interpretability through feature sparsity, on survival datasets. The final procedure is efficient, selects useful covariates and outperforms existing CoxPH approaches.
Lymph Node Graph Neural Networks for Cancer Metastasis Prediction
Jun 03, 2021



Abstract:Predicting outcomes, such as survival or metastasis for individual cancer patients is a crucial component of precision oncology. Machine learning (ML) offers a promising way to exploit rich multi-modal data, including clinical information and imaging to learn predictors of disease trajectory and help inform clinical decision making. In this paper, we present a novel graph-based approach to incorporate imaging characteristics of existing cancer spread to local lymph nodes (LNs) as well as their connectivity patterns in a prognostic ML model. We trained an edge-gated Graph Convolutional Network (Gated-GCN) to accurately predict the risk of distant metastasis (DM) by propagating information across the LN graph with the aid of soft edge attention mechanism. In a cohort of 1570 head and neck cancer patients, the Gated-GCN achieves AUROC of 0.757 for 2-year DM classification and $C$-index of 0.725 for lifetime DM risk prediction, outperforming current prognostic factors as well as previous approaches based on aggregated LN features. We also explored the importance of graph structure and individual lymph nodes through ablation experiments and interpretability studies, highlighting the importance of considering individual LN characteristics as well as the relationships between regions of cancer spread.
A Machine Learning Challenge for Prognostic Modelling in Head and Neck Cancer Using Multi-modal Data
Jan 28, 2021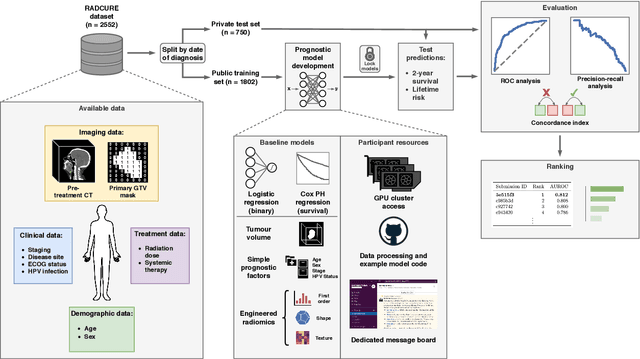

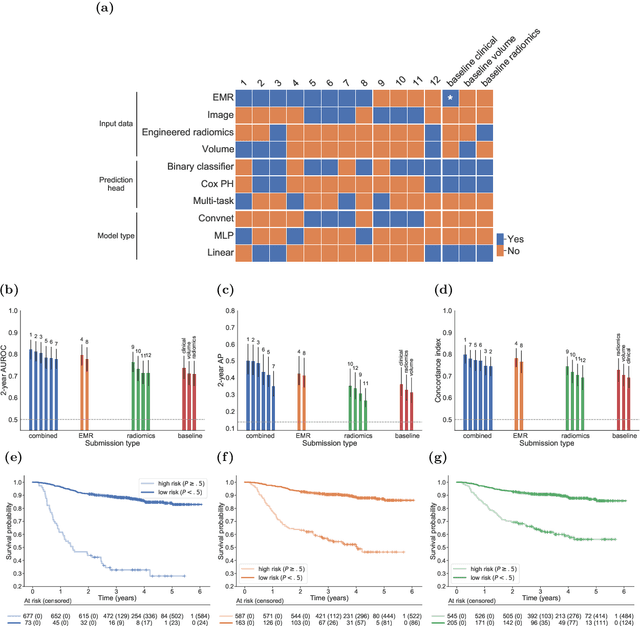
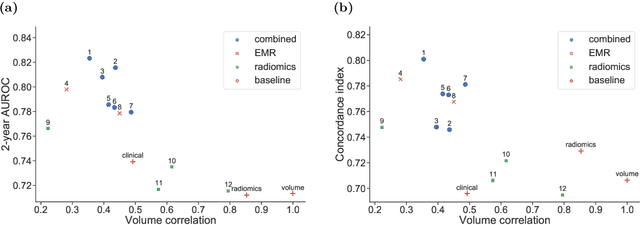
Abstract:Accurate prognosis for an individual patient is a key component of precision oncology. Recent advances in machine learning have enabled the development of models using a wider range of data, including imaging. Radiomics aims to extract quantitative predictive and prognostic biomarkers from routine medical imaging, but evidence for computed tomography radiomics for prognosis remains inconclusive. We have conducted an institutional machine learning challenge to develop an accurate model for overall survival prediction in head and neck cancer using clinical data etxracted from electronic medical records and pre-treatment radiological images, as well as to evaluate the true added benefit of radiomics for head and neck cancer prognosis. Using a large, retrospective dataset of 2,552 patients and a rigorous evaluation framework, we compared 12 different submissions using imaging and clinical data, separately or in combination. The winning approach used non-linear, multitask learning on clinical data and tumour volume, achieving high prognostic accuracy for 2-year and lifetime survival prediction and outperforming models relying on clinical data only, engineered radiomics and deep learning. Combining all submissions in an ensemble model resulted in improved accuracy, with the highest gain from a image-based deep learning model. Our results show the potential of machine learning and simple, informative prognostic factors in combination with large datasets as a tool to guide personalized cancer care.
Deep-CR MTLR: a Multi-Modal Approach for Cancer Survival Prediction with Competing Risks
Dec 10, 2020


Abstract:Accurate survival prediction is crucial for development of precision cancer medicine, creating the need for new sources of prognostic information. Recently, there has been significant interest in exploiting routinely collected clinical and medical imaging data to discover new prognostic markers in multiple cancer types. However, most of the previous studies focus on individual data modalities alone and do not make use of recent advances in machine learning for survival prediction. We present Deep-CR MTLR -- a novel machine learning approach for accurate cancer survival prediction from multi-modal clinical and imaging data in the presence of competing risks based on neural networks and an extension of the multi-task logistic regression framework. We demonstrate improved prognostic performance of the multi-modal approach over single modality predictors in a cohort of 2552 head and neck cancer patients, particularly for cancer specific survival, where our approach achieves 2-year AUROC of 0.774 and $C$-index of 0.788.
Learning across label confidence distributions using Filtered Transfer Learning
Jun 03, 2020



Abstract:Performance of neural network models relies on the availability of large datasets with minimal levels of uncertainty. Transfer Learning (TL) models have been proposed to resolve the issue of small dataset size by letting the model train on a bigger, task-related reference dataset and then fine-tune on a smaller, task-specific dataset. In this work, we apply a transfer learning approach to improve predictive power in noisy data systems with large variable confidence datasets. We propose a deep neural network method called Filtered Transfer Learning (FTL) that defines multiple tiers of data confidence as separate tasks in a transfer learning setting. The deep neural network is fine-tuned in a hierarchical process by iteratively removing (filtering) data points with lower label confidence, and retraining. In this report we use FTL for predicting the interaction of drugs and proteins. We demonstrate that using FTL to learn stepwise, across the label confidence distribution, results in higher performance compared to deep neural network models trained on a single confidence range. We anticipate that this approach will enable the machine learning community to benefit from large datasets with uncertain labels in fields such as biology and medicine.
CovidCTNet: An Open-Source Deep Learning Approach to Identify Covid-19 Using CT Image
May 16, 2020Abstract:Coronavirus disease 2019 (Covid-19) is highly contagious with limited treatment options. Early and accurate diagnosis of Covid-19 is crucial in reducing the spread of the disease and its accompanied mortality. Currently, detection by reverse transcriptase polymerase chain reaction (RT-PCR) is the gold standard of outpatient and inpatient detection of Covid-19. RT-PCR is a rapid method, however, its accuracy in detection is only ~70-75%. Another approved strategy is computed tomography (CT) imaging. CT imaging has a much higher sensitivity of ~80-98%, but similar accuracy of 70%. To enhance the accuracy of CT imaging detection, we developed an open-source set of algorithms called CovidCTNet that successfully differentiates Covid-19 from community-acquired pneumonia (CAP) and other lung diseases. CovidCTNet increases the accuracy of CT imaging detection to 90% compared to radiologists (70%). The model is designed to work with heterogeneous and small sample sizes independent of the CT imaging hardware. In order to facilitate the detection of Covid-19 globally and assist radiologists and physicians in the screening process, we are releasing all algorithms and parametric details in an open-source format. Open-source sharing of our CovidCTNet enables developers to rapidly improve and optimize services, while preserving user privacy and data ownership.
Reducing Adversarial Example Transferability Using Gradient Regularization
Apr 16, 2019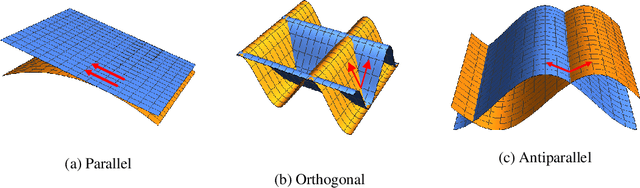



Abstract:Deep learning algorithms have increasingly been shown to lack robustness to simple adversarial examples (AdvX). An equally troubling observation is that these adversarial examples transfer between different architectures trained on different datasets. We investigate the transferability of adversarial examples between models using the angle between the input-output Jacobians of different models. To demonstrate the relevance of this approach, we perform case studies that involve jointly training pairs of models. These case studies empirically justify the theoretical intuitions for why the angle between gradients is a fundamental quantity in AdvX transferability. Furthermore, we consider the asymmetry of AdvX transferability between two models of the same architecture and explain it in terms of differences in gradient norms between the models. Lastly, we provide a simple modification to existing training setups that reduces transferability of adversarial examples between pairs of models.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge