Chun-Hao Chang
Data-Efficient and Interpretable Tabular Anomaly Detection
Mar 03, 2022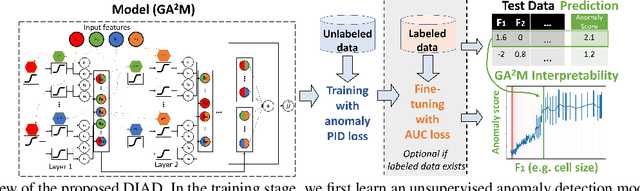
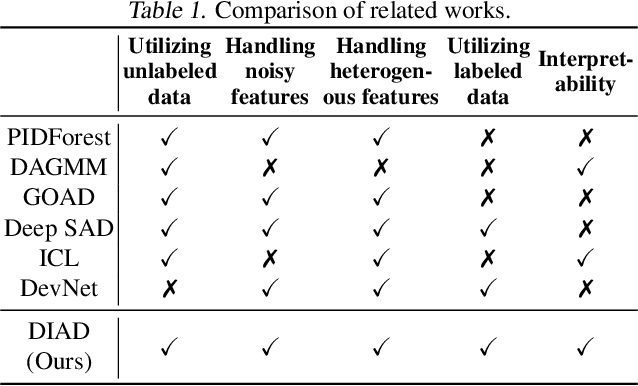
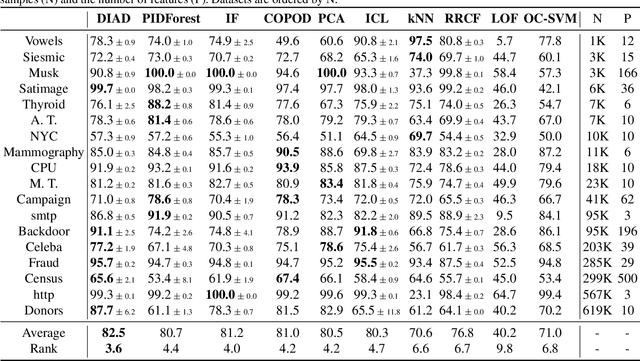
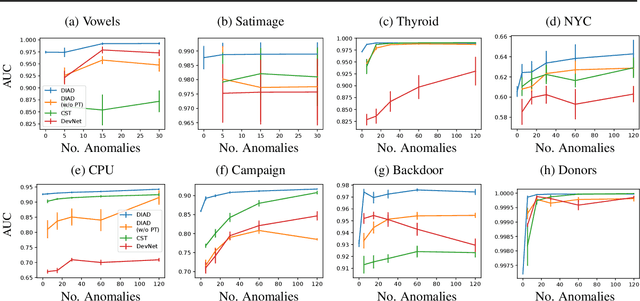
Abstract:Anomaly detection (AD) plays an important role in numerous applications. We focus on two understudied aspects of AD that are critical for integration into real-world applications. First, most AD methods cannot incorporate labeled data that are often available in practice in small quantities and can be crucial to achieve high AD accuracy. Second, most AD methods are not interpretable, a bottleneck that prevents stakeholders from understanding the reason behind the anomalies. In this paper, we propose a novel AD framework that adapts a white-box model class, Generalized Additive Models, to detect anomalies using a partial identification objective which naturally handles noisy or heterogeneous features. In addition, the proposed framework, DIAD, can incorporate a small amount of labeled data to further boost anomaly detection performances in semi-supervised settings. We demonstrate the superiority of our framework compared to previous work in both unsupervised and semi-supervised settings using diverse tabular datasets. For example, under 5 labeled anomalies DIAD improves from 86.2\% to 89.4\% AUC by learning AD from unlabeled data. We also present insightful interpretations that explain why DIAD deems certain samples as anomalies.
Extracting Clinician's Goals by What-if Interpretable Modeling
Oct 28, 2021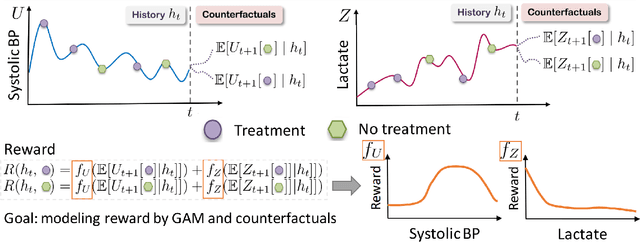
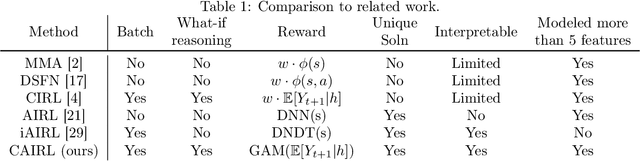
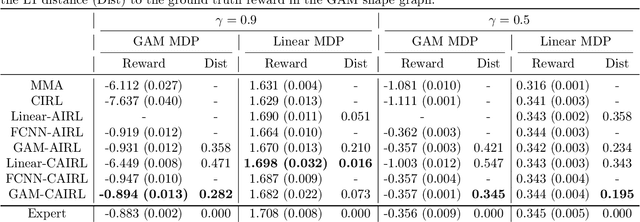
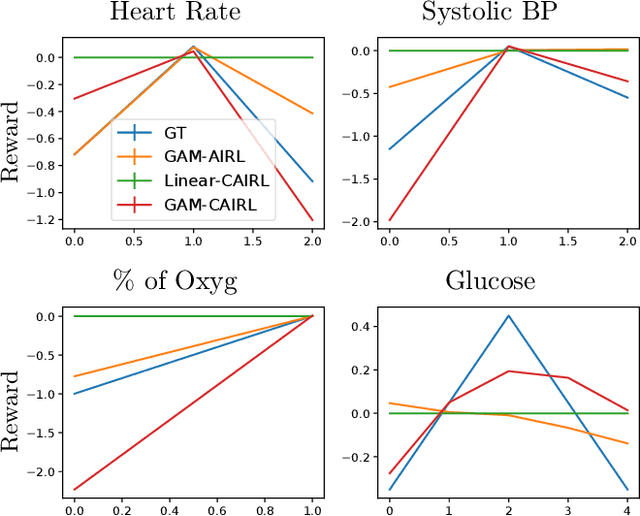
Abstract:Although reinforcement learning (RL) has tremendous success in many fields, applying RL to real-world settings such as healthcare is challenging when the reward is hard to specify and no exploration is allowed. In this work, we focus on recovering clinicians' rewards in treating patients. We incorporate the what-if reasoning to explain clinician's actions based on future outcomes. We use generalized additive models (GAMs) - a class of accurate, interpretable models - to recover the reward. In both simulation and a real-world hospital dataset, we show our model outperforms baselines. Finally, our model's explanations match several clinical guidelines when treating patients while we found the previously-used linear model often contradicts them.
NODE-GAM: Neural Generalized Additive Model for Interpretable Deep Learning
Jun 03, 2021



Abstract:Deployment of machine learning models in real high-risk settings (e.g. healthcare) often depends not only on model's accuracy but also on its fairness, robustness and interpretability. Generalized Additive Models (GAMs) have a long history of use in these high-risk domains, but lack desirable features of deep learning such as differentiability and scalability. In this work, we propose a neural GAM (NODE-GAM) and neural GA$^2$M (NODE-GA$^2$M) that scale well to large datasets, while remaining interpretable and accurate. We show that our proposed models have comparable accuracy to other non-interpretable models, and outperform other GAMs on large datasets. We also show that our models are more accurate in self-supervised learning setting when access to labeled data is limited.
Towards Robust Classification Model by Counterfactual and Invariant Data Generation
Jun 03, 2021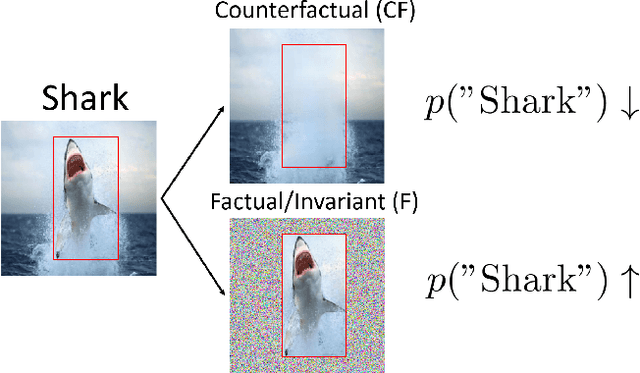
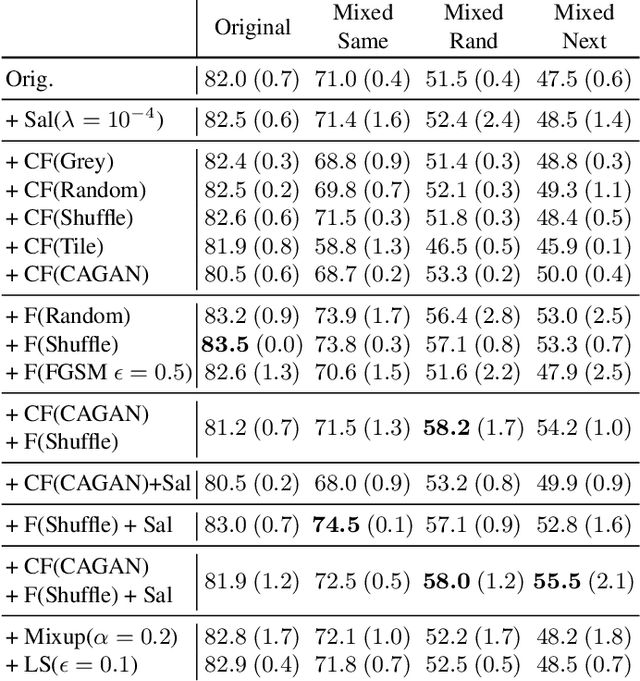

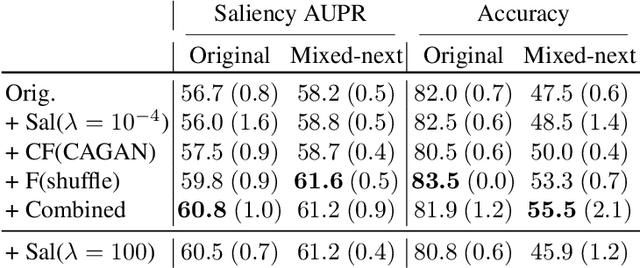
Abstract:Despite the success of machine learning applications in science, industry, and society in general, many approaches are known to be non-robust, often relying on spurious correlations to make predictions. Spuriousness occurs when some features correlate with labels but are not causal; relying on such features prevents models from generalizing to unseen environments where such correlations break. In this work, we focus on image classification and propose two data generation processes to reduce spuriousness. Given human annotations of the subset of the features responsible (causal) for the labels (e.g. bounding boxes), we modify this causal set to generate a surrogate image that no longer has the same label (i.e. a counterfactual image). We also alter non-causal features to generate images still recognized as the original labels, which helps to learn a model invariant to these features. In several challenging datasets, our data generations outperform state-of-the-art methods in accuracy when spurious correlations break, and increase the saliency focus on causal features providing better explanations.
How Interpretable and Trustworthy are GAMs?
Jun 11, 2020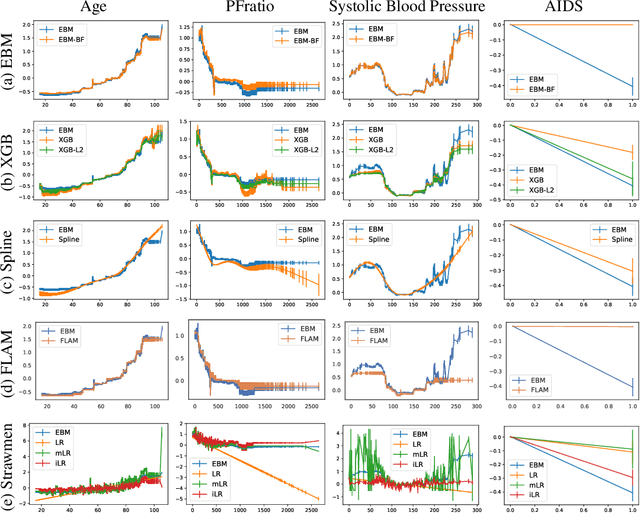

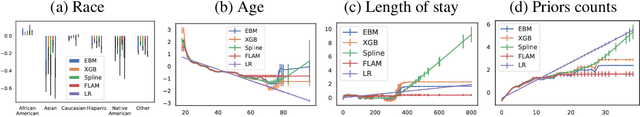

Abstract:Generalized additive models (GAMs) have become a leading model class for data bias discovery and model auditing. However, there are a variety of algorithms for training GAMs, and these do not always learn the same things. Statisticians originally used splines to train GAMs, but more recently GAMs are being trained with boosted decision trees. It is unclear which GAM model(s) to believe, particularly when their explanations are contradictory. In this paper, we investigate a variety of different GAM algorithms both qualitatively and quantitatively on real and simulated datasets. Our results suggest that inductive bias plays a crucial role in model explanations and tree-based GAMs are to be recommended for the kinds of problems and dataset sizes we worked with.
Purifying Interaction Effects with the Functional ANOVA: An Efficient Algorithm for Recovering Identifiable Additive Models
Nov 12, 2019



Abstract:Recent methods for training generalized additive models (GAMs) with pairwise interactions achieve state-of-the-art accuracy on a variety of datasets. Adding interactions to GAMs, however, introduces an identifiability problem: effects can be freely moved between main effects and interaction effects without changing the model predictions. In some cases, this can lead to contradictory interpretations of the same underlying function. This is a critical problem because a central motivation of GAMs is model interpretability. In this paper, we use the Functional ANOVA decomposition to uniquely define interaction effects and thus produce identifiable additive models with purified interactions. To compute this decomposition, we present a fast, exact, mass-moving algorithm that transforms any piecewise-constant function (such as a tree-based model) into a purified, canonical representation. We apply this algorithm to several datasets and show large disparity, including contradictions, between the apparent and the purified effects.
Dynamic Measurement Scheduling for Event Forecasting using Deep RL
Jan 24, 2019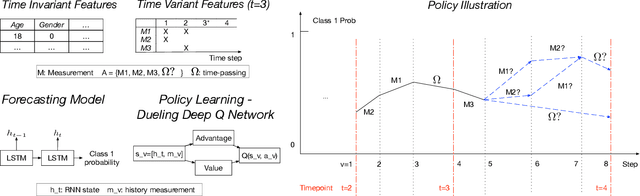
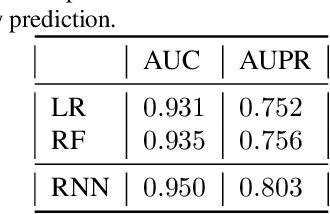
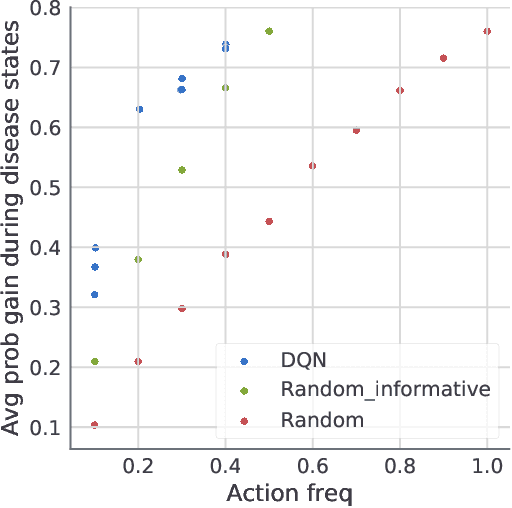
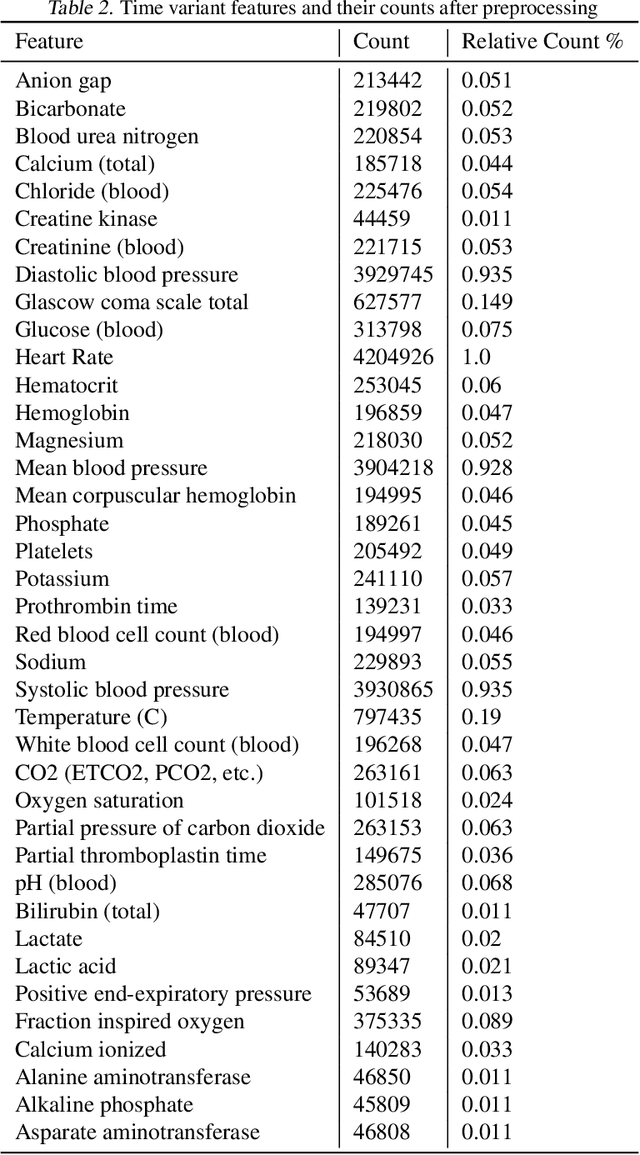
Abstract:Current clinical practice for monitoring patients' health follows either regular or heuristic-based lab test (e.g. blood test) scheduling. Such practice not only gives rise to redundant measurements accruing cost, but may even cause unnecessary patient discomfort. From the computational perspective, heuristic-based test scheduling might lead to reduced accuracy of clinical forecasting models. A data-driven measurement scheduling is likely to lead to both more accurate predictions and less measurement costs. We address the scheduling problem using deep reinforcement learning (RL) and propose a general and scalable framework to achieve high predictive gain and low measurement cost, by scheduling fewer, but strategically timed tests. Using simulations we show that our policy outperforms heuristic-based measurement scheduling with higher predictive gain and lower cost. We then learn a scheduling policy for mortality forecasting in the real-world clinical dataset (MIMIC3). Our policy decreases the total number of measurements by 31% without reducing the predictive performance, or improves 3 times more predictive gain with the same number of measurements using off-policy policy evaluation.
Dynamic Measurement Scheduling for Adverse Event Forecasting using Deep RL
Dec 01, 2018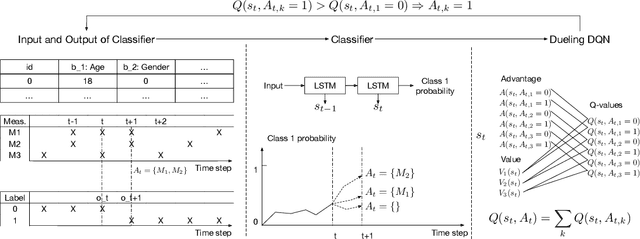
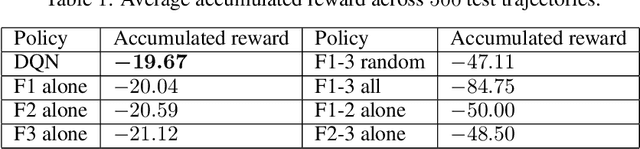


Abstract:Current clinical practice to monitor patients' health follows either regular or heuristic-based lab test (e.g. blood test) scheduling. Such practice not only gives rise to redundant measurements accruing cost, but may even lead to unnecessary patient discomfort. From the computational perspective, heuristic-based test scheduling might lead to reduced accuracy of clinical forecasting models. Computationally learning an optimal clinical test scheduling and measurement collection, is likely to lead to both, better predictive models and patient outcome improvement. We address the scheduling problem using deep reinforcement learning (RL) to achieve high predictive gain and low measurement cost, by scheduling fewer, but strategically timed tests. We first show that in the simulation our policy outperforms heuristic-based measurement scheduling with higher predictive gain or lower cost measured by accumulated reward. We then learn a scheduling policy for mortality forecasting in the real-world clinical dataset (MIMIC3), our learned policy is able to provide useful clinical insights. To our knowledge, this is the first RL application on multi-measurement scheduling problem in the clinical setting.
Explaining Image Classifiers by Counterfactual Generation
Oct 11, 2018



Abstract:When a black-box classifier processes an input to render a prediction, which input features are relevant and why? We propose to answer this question by efficiently marginalizing over the universe of plausible alternative values for a subset of features by conditioning a generative model of the input distribution on the remaining features. In contrast with recent approaches that compute alternative feature values ad-hoc --- generating counterfactual inputs far from the natural data distribution --- our model-agnostic method produces realistic explanations, generating plausible inputs that either preserve or alter the classification confidence. When applied to image classification, our method produces more compact and relevant per-feature saliency assignment, with fewer artifacts compared to previous methods.
Dropout Feature Ranking for Deep Learning Models
Mar 09, 2018



Abstract:Deep neural networks (DNNs) achieve state-of-the-art results in a variety of domains. Unfortunately, DNNs are notorious for their non-interpretability, and thus limit their applicability in hypothesis-driven domains such as biology and healthcare. Moreover, in the resource-constraint setting, it is critical to design tests relying on fewer more informative features leading to high accuracy performance within reasonable budget. We aim to close this gap by proposing a new general feature ranking method for deep learning. We show that our simple yet effective method performs on par or compares favorably to eight strawman, classical and deep-learning feature ranking methods in two simulations and five very different datasets on tasks ranging from classification to regression, in both static and time series scenarios. We also illustrate the use of our method on a drug response dataset and show that it identifies genes relevant to the drug-response.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge