Demian Wassermann
PARIETAL
Robust and highly scalable estimation of directional couplings from time-shifted signals
Jun 04, 2024
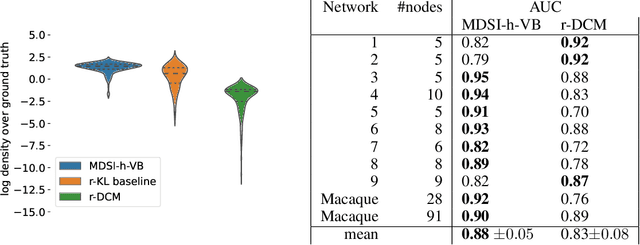


Abstract:The estimation of directed couplings between the nodes of a network from indirect measurements is a central methodological challenge in scientific fields such as neuroscience, systems biology and economics. Unfortunately, the problem is generally ill-posed due to the possible presence of unknown delays in the measurements. In this paper, we offer a solution of this problem by using a variational Bayes framework, where the uncertainty over the delays is marginalized in order to obtain conservative coupling estimates. To overcome the well-known overconfidence of classical variational methods, we use a hybrid-VI scheme where the (possibly flat or multimodal) posterior over the measurement parameters is estimated using a forward KL loss while the (nearly convex) conditional posterior over the couplings is estimated using the highly scalable gradient-based VI. In our ground-truth experiments, we show that the network provides reliable and conservative estimates of the couplings, greatly outperforming similar methods such as regression DCM.
PAVI: Plate-Amortized Variational Inference
Aug 30, 2023Abstract:Given observed data and a probabilistic generative model, Bayesian inference searches for the distribution of the model's parameters that could have yielded the data. Inference is challenging for large population studies where millions of measurements are performed over a cohort of hundreds of subjects, resulting in a massive parameter space. This large cardinality renders off-the-shelf Variational Inference (VI) computationally impractical. In this work, we design structured VI families that efficiently tackle large population studies. Our main idea is to share the parameterization and learning across the different i.i.d. variables in a generative model, symbolized by the model's \textit{plates}. We name this concept \textit{plate amortization}. Contrary to off-the-shelf stochastic VI, which slows down inference, plate amortization results in orders of magnitude faster to train variational distributions. Applied to large-scale hierarchical problems, PAVI yields expressive, parsimoniously parameterized VI with an affordable training time. This faster convergence effectively unlocks inference in those large regimes. We illustrate the practical utility of PAVI through a challenging Neuroimaging example featuring 400 million latent parameters, demonstrating a significant step towards scalable and expressive Variational Inference.
Scalable Query Answering under Uncertainty to Neuroscientific Ontological Knowledge: The NeuroLang Approach
Feb 23, 2022
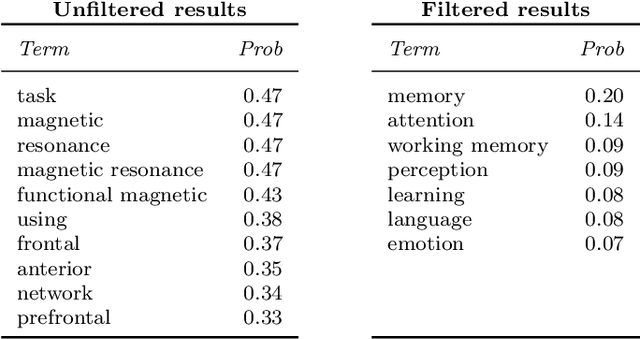
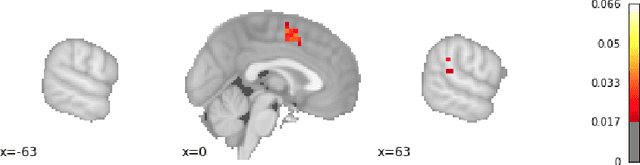
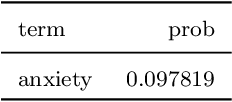
Abstract:Researchers in neuroscience have a growing number of datasets available to study the brain, which is made possible by recent technological advances. Given the extent to which the brain has been studied, there is also available ontological knowledge encoding the current state of the art regarding its different areas, activation patterns, key words associated with studies, etc. Furthermore, there is an inherent uncertainty associated with brain scans arising from the mapping between voxels -- 3D pixels -- and actual points in different individual brains. Unfortunately, there is currently no unifying framework for accessing such collections of rich heterogeneous data under uncertainty, making it necessary for researchers to rely on ad hoc tools. In particular, one major weakness of current tools that attempt to address this kind of task is that only very limited propositional query languages have been developed. In this paper, we present NeuroLang, an ontology language with existential rules, probabilistic uncertainty, and built-in mechanisms to guarantee tractable query answering over very large datasets. After presenting the language and its general query answering architecture, we discuss real-world use cases showing how NeuroLang can be applied to practical scenarios for which current tools are inadequate.
Inverting brain grey matter models with likelihood-free inference: a tool for trustable cytoarchitecture measurements
Nov 15, 2021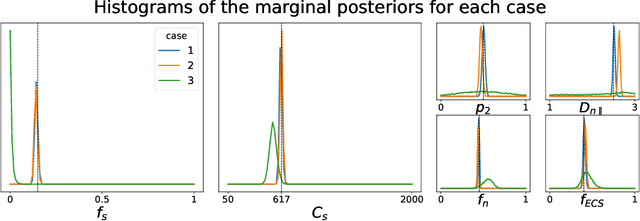
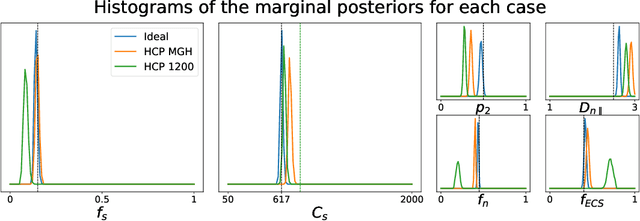
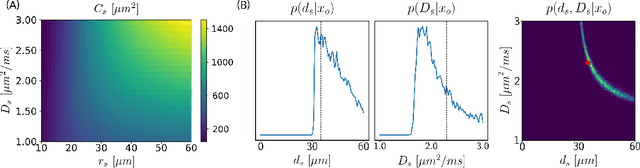
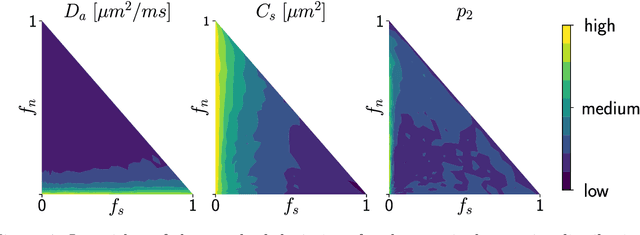
Abstract:Effective characterisation of the brain grey matter cytoarchitecture with quantitative sensitivity to soma density and volume remains an unsolved challenge in diffusion MRI (dMRI). Solving the problem of relating the dMRI signal with cytoarchitectural characteristics calls for the definition of a mathematical model that describes brain tissue via a handful of physiologically-relevant parameters and an algorithm for inverting the model. To address this issue, we propose a new forward model, specifically a new system of equations, requiring a few relatively sparse b-shells. We then apply modern tools from Bayesian analysis known as likelihood-free inference (LFI) to invert our proposed model. As opposed to other approaches from the literature, our algorithm yields not only an estimation of the parameter vector $\theta$ that best describes a given observed data point $x_0$, but also a full posterior distribution $p(\theta|x_0)$ over the parameter space. This enables a richer description of the model inversion, providing indicators such as credible intervals for the estimated parameters and a complete characterization of the parameter regions where the model may present indeterminacies. We approximate the posterior distribution using deep neural density estimators, known as normalizing flows, and fit them using a set of repeated simulations from the forward model. We validate our approach on simulations using dmipy and then apply the whole pipeline on two publicly available datasets.
ADAVI: Automatic Dual Amortized Variational Inference Applied To Pyramidal Bayesian Models
Jun 23, 2021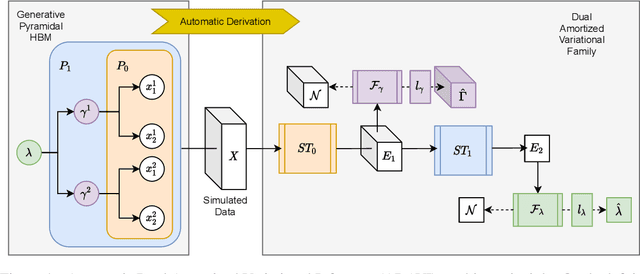
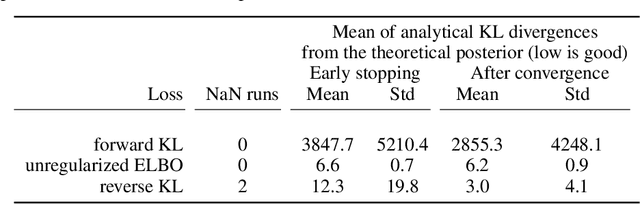
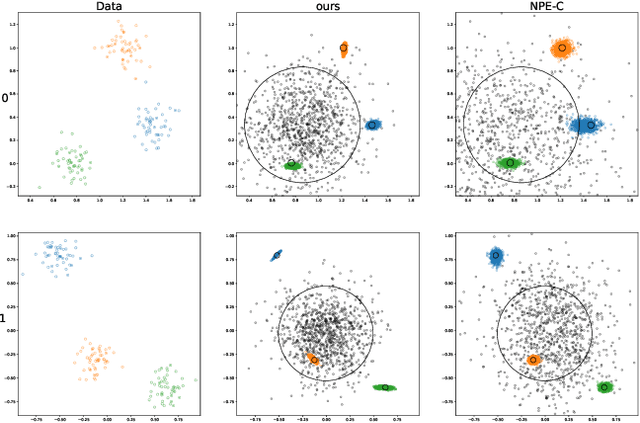
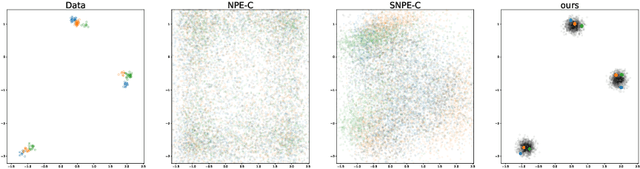
Abstract:Frequently, population studies feature pyramidally-organized data represented using Hierarchical Bayesian Models (HBM) enriched with plates. These models can become prohibitively large in settings such as neuroimaging, where a sample is composed of a functional MRI signal measured on 64 thousand brain locations, across 4 measurement sessions, and at least tens of subjects. Even a reduced example on a specific cortical region of 300 brain locations features around 1 million parameters, hampering the usage of modern density estimation techniques such as Simulation-Based Inference (SBI). To infer parameter posterior distributions in this challenging class of problems, we designed a novel methodology that automatically produces a variational family dual to a target HBM. This variatonal family, represented as a neural network, consists in the combination of an attention-based hierarchical encoder feeding summary statistics to a set of normalizing flows. Our automatically-derived neural network exploits exchangeability in the plate-enriched HBM and factorizes its parameter space. The resulting architecture reduces by orders of magnitude its parameterization with respect to that of a typical SBI representation, while maintaining expressivity. Our method performs inference on the specified HBM in an amortized setup: once trained, it can readily be applied to a new data sample to compute the parameters' full posterior. We demonstrate the capability of our method on simulated data, as well as a challenging high-dimensional brain parcellation experiment. We also open up several questions that lie at the intersection between SBI techniques and structured Variational Inference.
Complex Coordinate-Based Meta-Analysis with Probabilistic Programming
Dec 02, 2020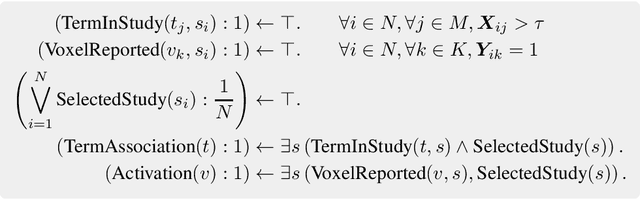
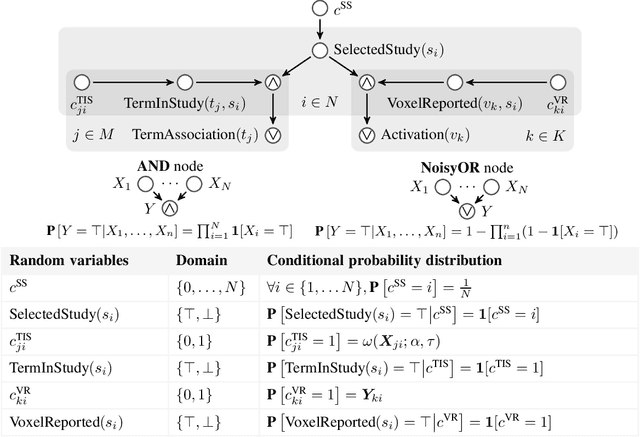

Abstract:With the growing number of published functional magnetic resonance imaging (fMRI) studies, meta-analysis databases and models have become an integral part of brain mapping research. Coordinate-based meta-analysis (CBMA) databases are built by automatically extracting both coordinates of reported peak activations and term associations using natural language processing (NLP) techniques. Solving term-based queries on these databases make it possible to obtain statistical maps of the brain related to specific cognitive processes. However, with tools like Neurosynth, only singleterm queries lead to statistically reliable results. When solving richer queries, too few studies from the database contribute to the statistical estimations. We design a probabilistic domain-specific language (DSL) standing on Datalog and one of its probabilistic extensions, CP-Logic, for expressing and solving rich logic-based queries. We encode a CBMA database into a probabilistic program. Using the joint distribution of its Bayesian network translation, we show that solutions of queries on this program compute the right probability distributions of voxel activations. We explain how recent lifted query processing algorithms make it possible to scale to the size of large neuroimaging data, where state of the art knowledge compilation (KC) techniques fail to solve queries fast enough for practical applications. Finally, we introduce a method for relating studies to terms probabilistically, leading to better solutions for conjunctive queries on smaller databases. We demonstrate results for two-term conjunctive queries, both on simulated meta-analysis databases and on the widely-used Neurosynth database.
PEP: Parameter Ensembling by Perturbation
Oct 24, 2020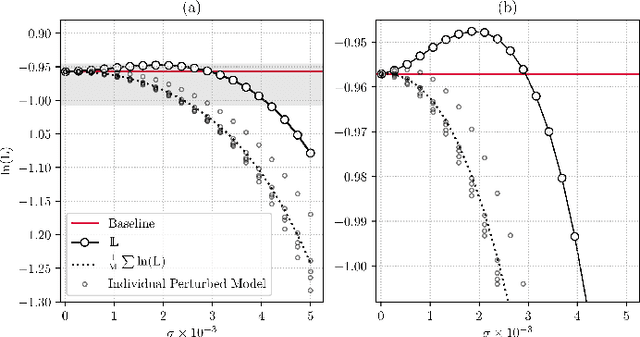
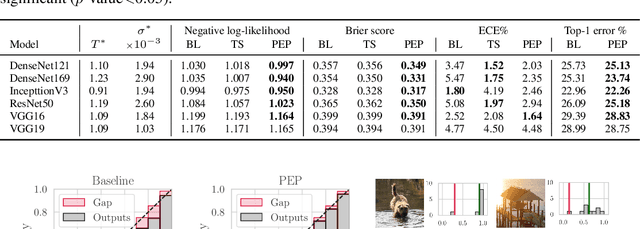
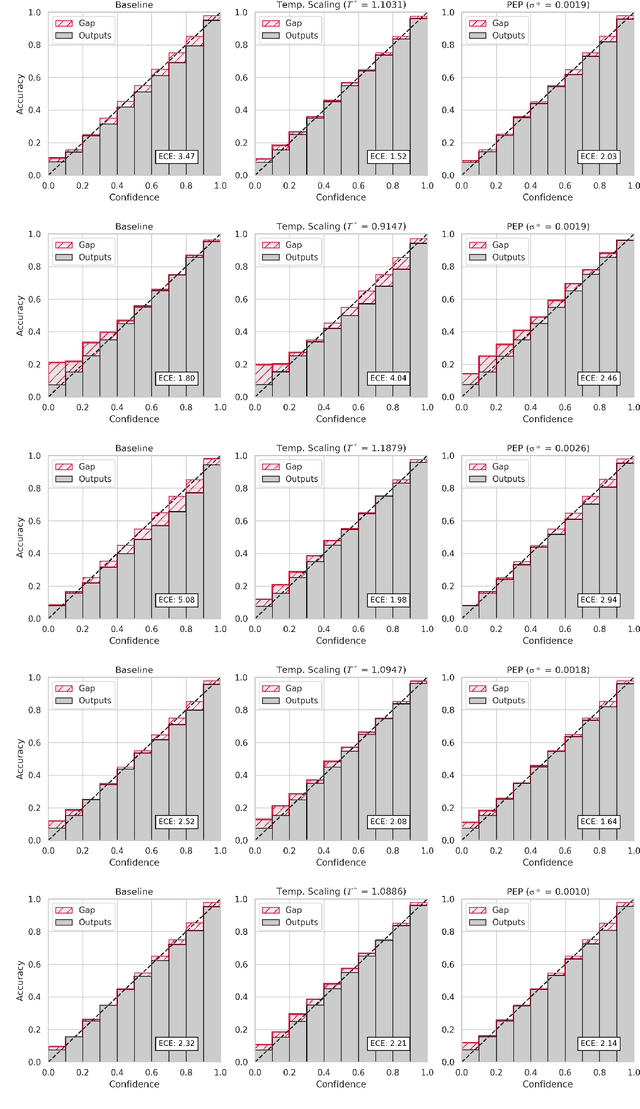
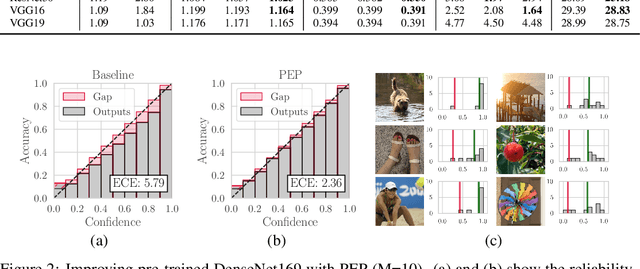
Abstract:Ensembling is now recognized as an effective approach for increasing the predictive performance and calibration of deep networks. We introduce a new approach, Parameter Ensembling by Perturbation (PEP), that constructs an ensemble of parameter values as random perturbations of the optimal parameter set from training by a Gaussian with a single variance parameter. The variance is chosen to maximize the log-likelihood of the ensemble average ($\mathbb{L}$) on the validation data set. Empirically, and perhaps surprisingly, $\mathbb{L}$ has a well-defined maximum as the variance grows from zero (which corresponds to the baseline model). Conveniently, calibration level of predictions also tends to grow favorably until the peak of $\mathbb{L}$ is reached. In most experiments, PEP provides a small improvement in performance, and, in some cases, a substantial improvement in empirical calibration. We show that this "PEP effect" (the gain in log-likelihood) is related to the mean curvature of the likelihood function and the empirical Fisher information. Experiments on ImageNet pre-trained networks including ResNet, DenseNet, and Inception showed improved calibration and likelihood. We further observed a mild improvement in classification accuracy on these networks. Experiments on classification benchmarks such as MNIST and CIFAR-10 showed improved calibration and likelihood, as well as the relationship between the PEP effect and overfitting; this demonstrates that PEP can be used to probe the level of overfitting that occurred during training. In general, no special training procedure or network architecture is needed, and in the case of pre-trained networks, no additional training is needed.
Fine-grain atlases of functional modes for fMRI analysis
Mar 05, 2020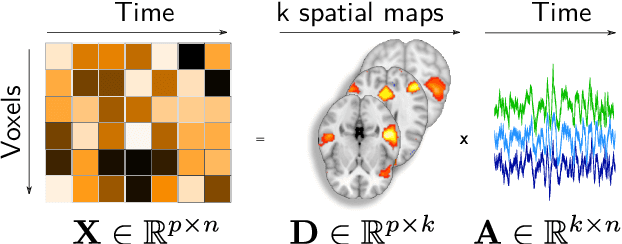
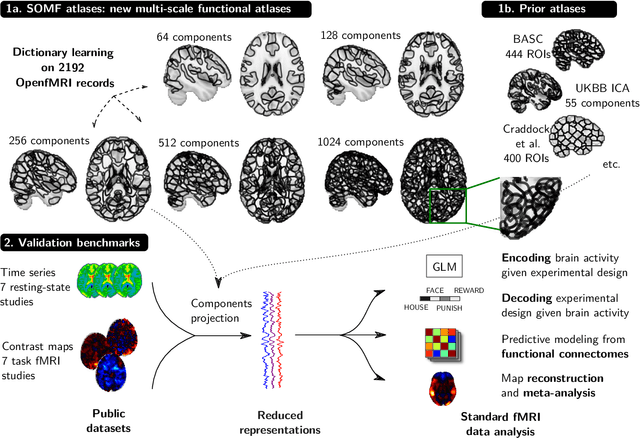
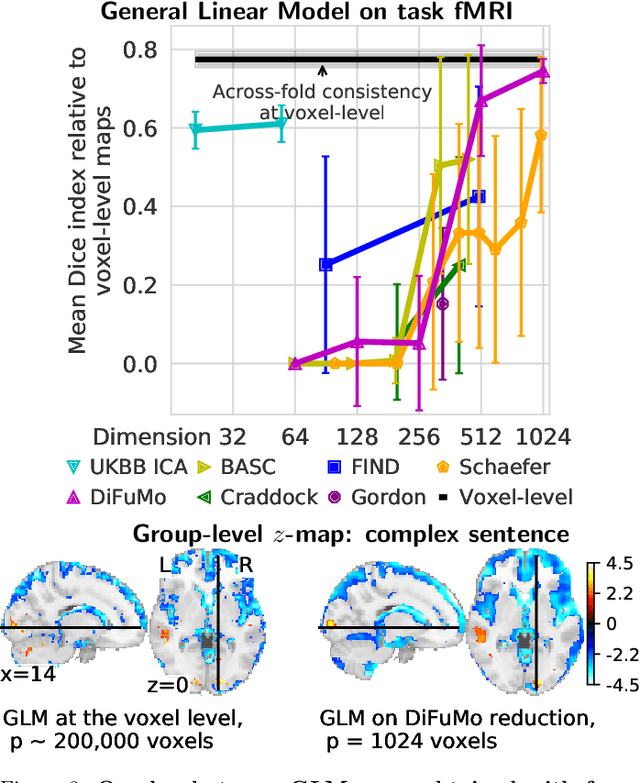
Abstract:Population imaging markedly increased the size of functional-imaging datasets, shedding new light on the neural basis of inter-individual differences. Analyzing these large data entails new scalability challenges, computational and statistical. For this reason, brain images are typically summarized in a few signals, for instance reducing voxel-level measures with brain atlases or functional modes. A good choice of the corresponding brain networks is important, as most data analyses start from these reduced signals. We contribute finely-resolved atlases of functional modes, comprising from 64 to 1024 networks. These dictionaries of functional modes (DiFuMo) are trained on millions of fMRI functional brain volumes of total size 2.4TB, spanned over 27 studies and many research groups. We demonstrate the benefits of extracting reduced signals on our fine-grain atlases for many classic functional data analysis pipelines: stimuli decoding from 12,334 brain responses, standard GLM analysis of fMRI across sessions and individuals, extraction of resting-state functional-connectomes biomarkers for 2,500 individuals, data compression and meta-analysis over more than 15,000 statistical maps. In each of these analysis scenarii, we compare the performance of our functional atlases with that of other popular references, and to a simple voxel-level analysis. Results highlight the importance of using high-dimensional "soft" functional atlases, to represent and analyse brain activity while capturing its functional gradients. Analyses on high-dimensional modes achieve similar statistical performance as at the voxel level, but with much reduced computational cost and higher interpretability. In addition to making them available, we provide meaningful names for these modes, based on their anatomical location. It will facilitate reporting of results.
Text to brain: predicting the spatial distribution of neuroimaging observations from text reports
Jun 28, 2018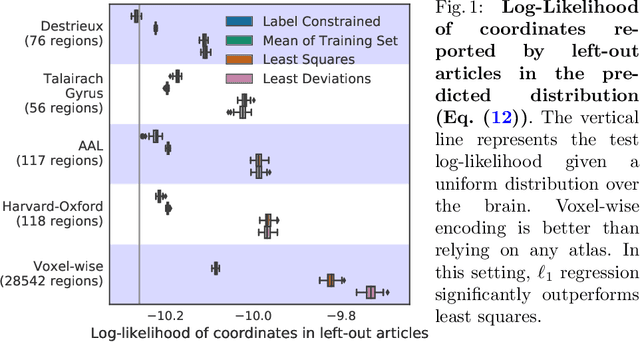
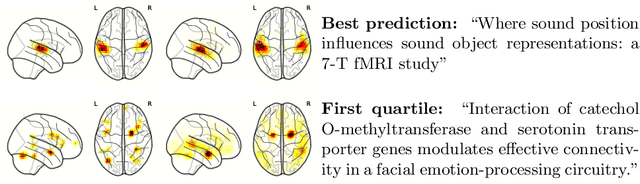
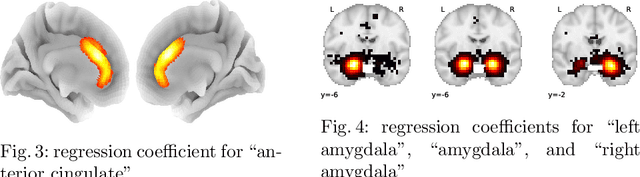
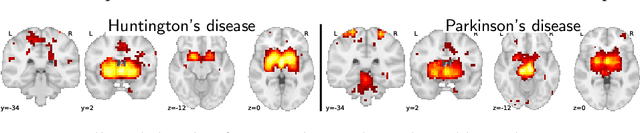
Abstract:Despite the digital nature of magnetic resonance imaging, the resulting observations are most frequently reported and stored in text documents. There is a trove of information untapped in medical health records, case reports, and medical publications. In this paper, we propose to mine brain medical publications to learn the spatial distribution associated with anatomical terms. The problem is formulated in terms of minimization of a risk on distributions which leads to a least-deviation cost function. An efficient algorithm in the dual then learns the mapping from documents to brain structures. Empirical results using coordinates extracted from the brain-imaging literature show that i) models must adapt to semantic variation in the terms used to describe a given anatomical structure, ii) voxel-wise parameterization leads to higher likelihood of locations reported in unseen documents, iii) least-deviation cost outperforms least-square. As a proof of concept for our method, we use our model of spatial distributions to predict the distribution of specific neurological conditions from text-only reports.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge