Jérôme Dockès
Inria
NeuroQuery: comprehensive meta-analysis of human brain mapping
Feb 21, 2020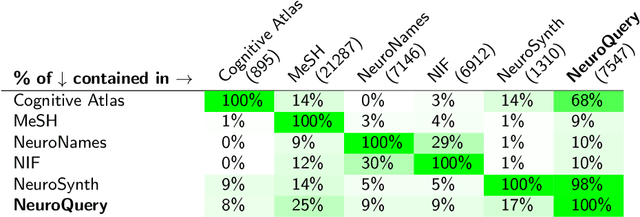
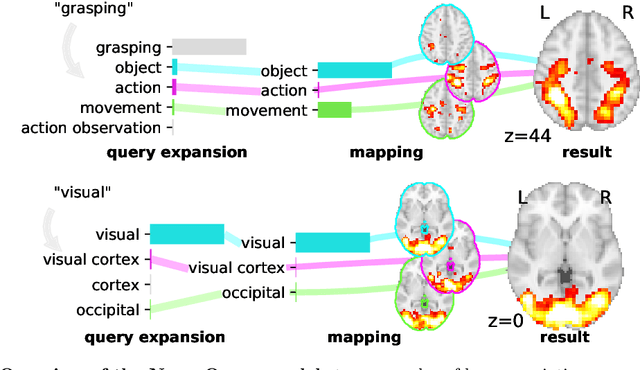
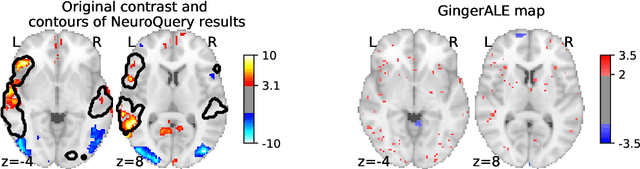
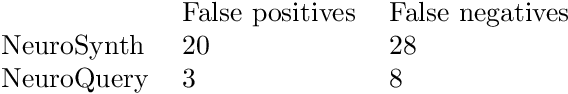
Abstract:Reaching a global view of brain organization requires assembling evidence on widely different mental processes and mechanisms. The variety of human neuroscience concepts and terminology poses a fundamental challenge to relating brain imaging results across the scientific literature. Existing meta-analysis methods perform statistical tests on sets of publications associated with a particular concept. Thus, large-scale meta-analyses only tackle single terms that occur frequently. We propose a new paradigm, focusing on prediction rather than inference. Our multivariate model predicts the spatial distribution of neurological observations, given text describing an experiment, cognitive process, or disease. This approach handles text of arbitrary length and terms that are too rare for standard meta-analysis. We capture the relationships and neural correlates of 7 547 neuroscience terms across 13 459 neuroimaging publications. The resulting meta-analytic tool, neuroquery.org, can ground hypothesis generation and data-analysis priors on a comprehensive view of published findings on the brain.
Text to brain: predicting the spatial distribution of neuroimaging observations from text reports
Jun 28, 2018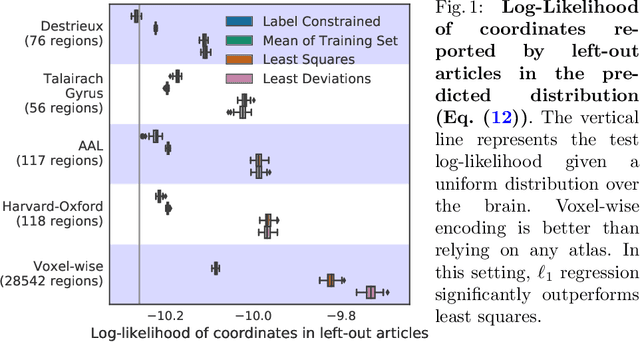
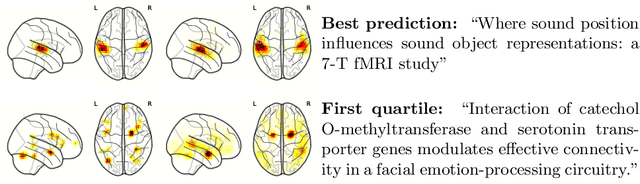
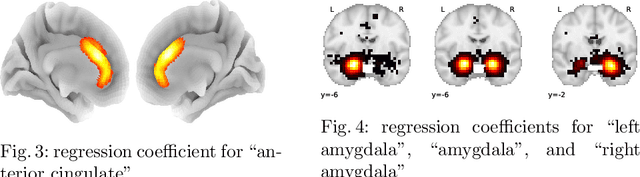
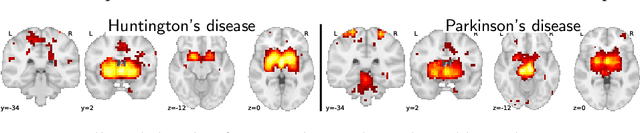
Abstract:Despite the digital nature of magnetic resonance imaging, the resulting observations are most frequently reported and stored in text documents. There is a trove of information untapped in medical health records, case reports, and medical publications. In this paper, we propose to mine brain medical publications to learn the spatial distribution associated with anatomical terms. The problem is formulated in terms of minimization of a risk on distributions which leads to a least-deviation cost function. An efficient algorithm in the dual then learns the mapping from documents to brain structures. Empirical results using coordinates extracted from the brain-imaging literature show that i) models must adapt to semantic variation in the terms used to describe a given anatomical structure, ii) voxel-wise parameterization leads to higher likelihood of locations reported in unseen documents, iii) least-deviation cost outperforms least-square. As a proof of concept for our method, we use our model of spatial distributions to predict the distribution of specific neurological conditions from text-only reports.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge