Tal Yarkoni
University of Texas
NeuroQuery: comprehensive meta-analysis of human brain mapping
Feb 21, 2020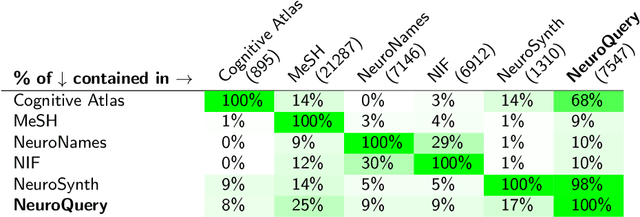
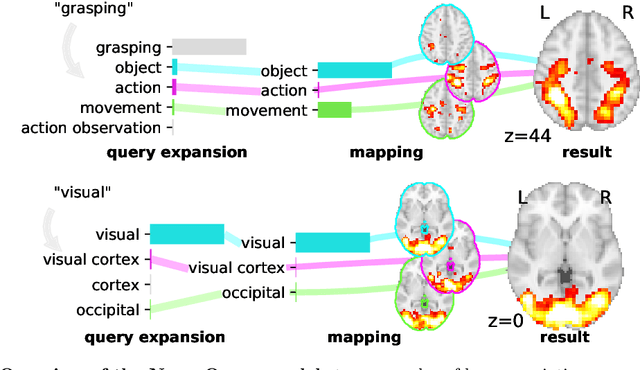
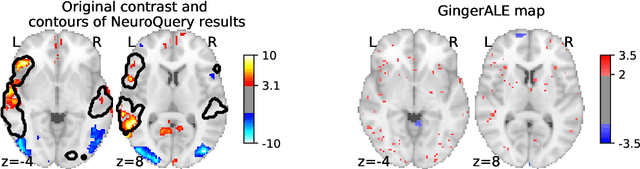
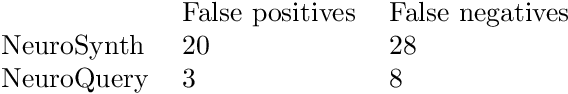
Abstract:Reaching a global view of brain organization requires assembling evidence on widely different mental processes and mechanisms. The variety of human neuroscience concepts and terminology poses a fundamental challenge to relating brain imaging results across the scientific literature. Existing meta-analysis methods perform statistical tests on sets of publications associated with a particular concept. Thus, large-scale meta-analyses only tackle single terms that occur frequently. We propose a new paradigm, focusing on prediction rather than inference. Our multivariate model predicts the spatial distribution of neurological observations, given text describing an experiment, cognitive process, or disease. This approach handles text of arbitrary length and terms that are too rare for standard meta-analysis. We capture the relationships and neural correlates of 7 547 neuroscience terms across 13 459 neuroimaging publications. The resulting meta-analytic tool, neuroquery.org, can ground hypothesis generation and data-analysis priors on a comprehensive view of published findings on the brain.
Developing a comprehensive framework for multimodal feature extraction
Feb 20, 2017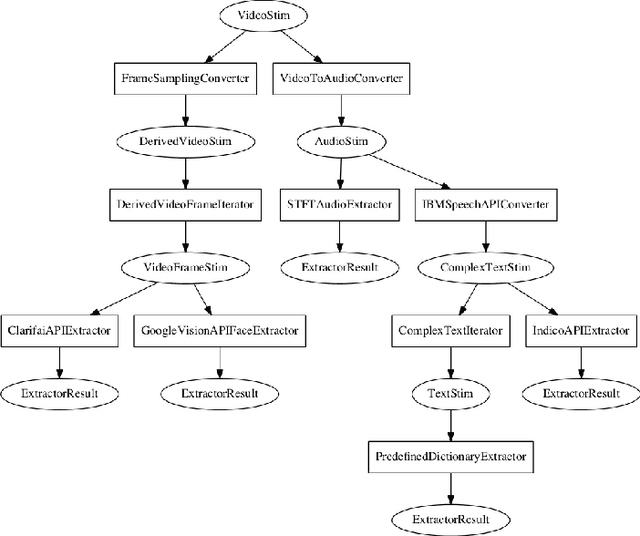
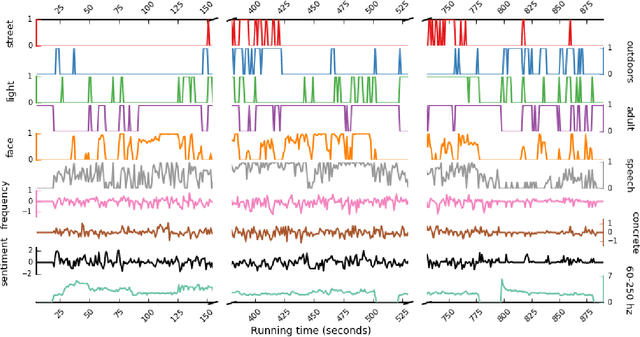
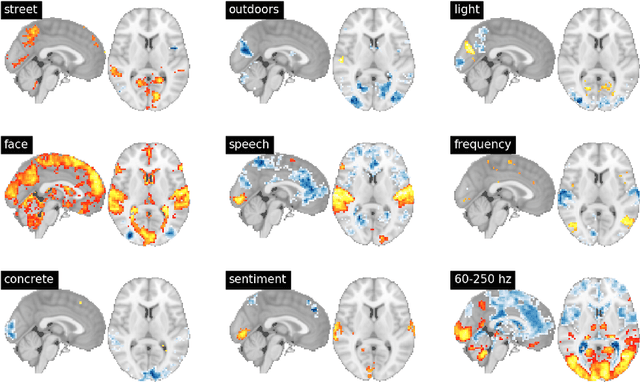
Abstract:Feature extraction is a critical component of many applied data science workflows. In recent years, rapid advances in artificial intelligence and machine learning have led to an explosion of feature extraction tools and services that allow data scientists to cheaply and effectively annotate their data along a vast array of dimensions---ranging from detecting faces in images to analyzing the sentiment expressed in coherent text. Unfortunately, the proliferation of powerful feature extraction services has been mirrored by a corresponding expansion in the number of distinct interfaces to feature extraction services. In a world where nearly every new service has its own API, documentation, and/or client library, data scientists who need to combine diverse features obtained from multiple sources are often forced to write and maintain ever more elaborate feature extraction pipelines. To address this challenge, we introduce a new open-source framework for comprehensive multimodal feature extraction. Pliers is an open-source Python package that supports standardized annotation of diverse data types (video, images, audio, and text), and is expressly with both ease-of-use and extensibility in mind. Users can apply a wide range of pre-existing feature extraction tools to their data in just a few lines of Python code, and can also easily add their own custom extractors by writing modular classes. A graph-based API enables rapid development of complex feature extraction pipelines that output results in a single, standardized format. We describe the package's architecture, detail its major advantages over previous feature extraction toolboxes, and use a sample application to a large functional MRI dataset to illustrate how pliers can significantly reduce the time and effort required to construct sophisticated feature extraction workflows while increasing code clarity and maintainability.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge