ADAVI: Automatic Dual Amortized Variational Inference Applied To Pyramidal Bayesian Models
Paper and Code
Jun 23, 2021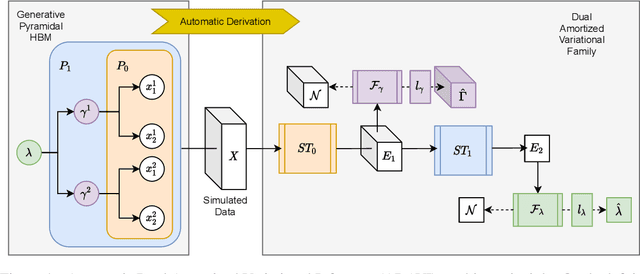
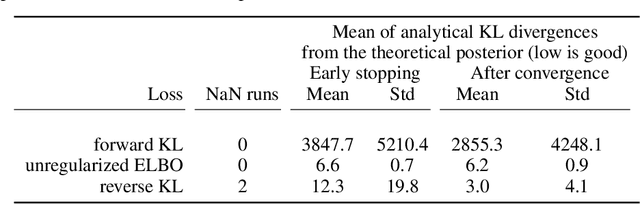
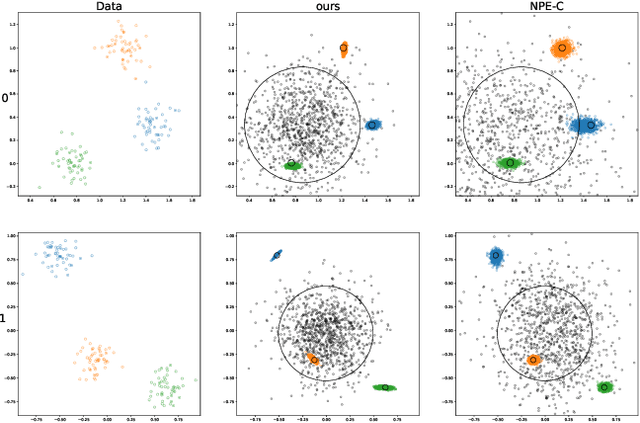
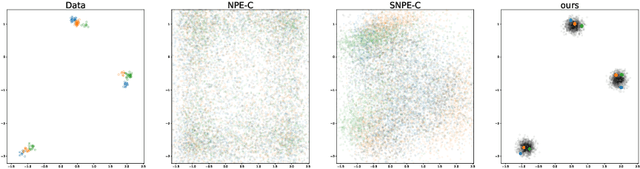
Frequently, population studies feature pyramidally-organized data represented using Hierarchical Bayesian Models (HBM) enriched with plates. These models can become prohibitively large in settings such as neuroimaging, where a sample is composed of a functional MRI signal measured on 64 thousand brain locations, across 4 measurement sessions, and at least tens of subjects. Even a reduced example on a specific cortical region of 300 brain locations features around 1 million parameters, hampering the usage of modern density estimation techniques such as Simulation-Based Inference (SBI). To infer parameter posterior distributions in this challenging class of problems, we designed a novel methodology that automatically produces a variational family dual to a target HBM. This variatonal family, represented as a neural network, consists in the combination of an attention-based hierarchical encoder feeding summary statistics to a set of normalizing flows. Our automatically-derived neural network exploits exchangeability in the plate-enriched HBM and factorizes its parameter space. The resulting architecture reduces by orders of magnitude its parameterization with respect to that of a typical SBI representation, while maintaining expressivity. Our method performs inference on the specified HBM in an amortized setup: once trained, it can readily be applied to a new data sample to compute the parameters' full posterior. We demonstrate the capability of our method on simulated data, as well as a challenging high-dimensional brain parcellation experiment. We also open up several questions that lie at the intersection between SBI techniques and structured Variational Inference.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge