Ricardo Rodriguez
UNS
LEARNER: Learning Granular Labels from Coarse Labels using Contrastive Learning
Nov 02, 2024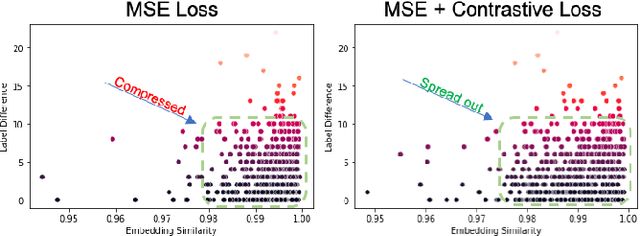

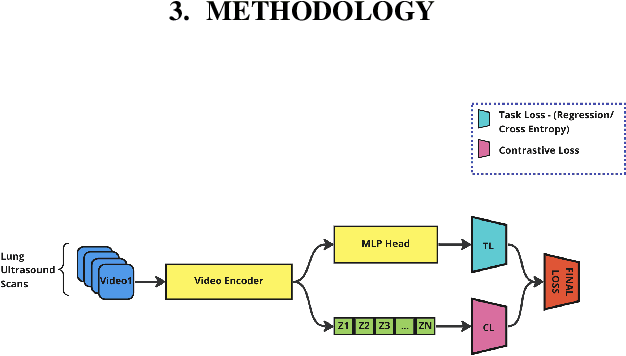
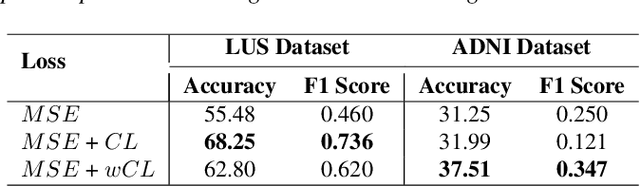
Abstract:A crucial question in active patient care is determining if a treatment is having the desired effect, especially when changes are subtle over short periods. We propose using inter-patient data to train models that can learn to detect these fine-grained changes within a single patient. Specifically, can a model trained on multi-patient scans predict subtle changes in an individual patient's scans? Recent years have seen increasing use of deep learning (DL) in predicting diseases using biomedical imaging, such as predicting COVID-19 severity using lung ultrasound (LUS) data. While extensive literature exists on successful applications of DL systems when well-annotated large-scale datasets are available, it is quite difficult to collect a large corpus of personalized datasets for an individual. In this work, we investigate the ability of recent computer vision models to learn fine-grained differences while being trained on data showing larger differences. We evaluate on an in-house LUS dataset and a public ADNI brain MRI dataset. We find that models pre-trained on clips from multiple patients can better predict fine-grained differences in scans from a single patient by employing contrastive learning.
Scalable Query Answering under Uncertainty to Neuroscientific Ontological Knowledge: The NeuroLang Approach
Feb 23, 2022
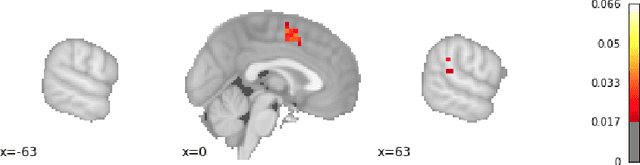
Abstract:Researchers in neuroscience have a growing number of datasets available to study the brain, which is made possible by recent technological advances. Given the extent to which the brain has been studied, there is also available ontological knowledge encoding the current state of the art regarding its different areas, activation patterns, key words associated with studies, etc. Furthermore, there is an inherent uncertainty associated with brain scans arising from the mapping between voxels -- 3D pixels -- and actual points in different individual brains. Unfortunately, there is currently no unifying framework for accessing such collections of rich heterogeneous data under uncertainty, making it necessary for researchers to rely on ad hoc tools. In particular, one major weakness of current tools that attempt to address this kind of task is that only very limited propositional query languages have been developed. In this paper, we present NeuroLang, an ontology language with existential rules, probabilistic uncertainty, and built-in mechanisms to guarantee tractable query answering over very large datasets. After presenting the language and its general query answering architecture, we discuss real-world use cases showing how NeuroLang can be applied to practical scenarios for which current tools are inadequate.
Simplified Kripke semantics for K45-like Godel modal logics and its axiomatic extensions
May 13, 2021Abstract:In this paper, we provide simplified semantics for the logic K45(G), i.e. the many-valued Godel counterpart of the classical modal logic K45. More precisely, we characterize K45(G) as the set of valid formulae of the class of possibilistic Godel Kripke Frames <W,\pi> where W is a non-empty set of worlds and \pi: W \to [0, 1] is a possibility distribution on W.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge