Amita Krishnan
LEARNER: Learning Granular Labels from Coarse Labels using Contrastive Learning
Nov 02, 2024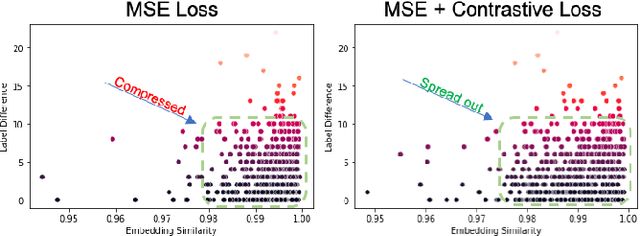

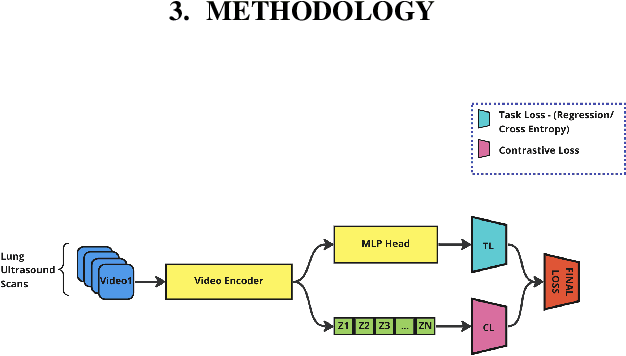
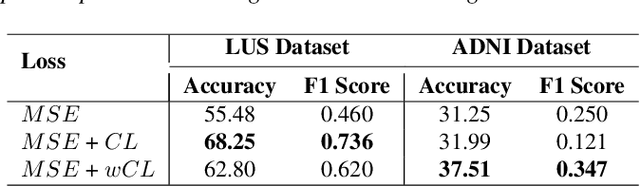
Abstract:A crucial question in active patient care is determining if a treatment is having the desired effect, especially when changes are subtle over short periods. We propose using inter-patient data to train models that can learn to detect these fine-grained changes within a single patient. Specifically, can a model trained on multi-patient scans predict subtle changes in an individual patient's scans? Recent years have seen increasing use of deep learning (DL) in predicting diseases using biomedical imaging, such as predicting COVID-19 severity using lung ultrasound (LUS) data. While extensive literature exists on successful applications of DL systems when well-annotated large-scale datasets are available, it is quite difficult to collect a large corpus of personalized datasets for an individual. In this work, we investigate the ability of recent computer vision models to learn fine-grained differences while being trained on data showing larger differences. We evaluate on an in-house LUS dataset and a public ADNI brain MRI dataset. We find that models pre-trained on clips from multiple patients can better predict fine-grained differences in scans from a single patient by employing contrastive learning.
Improving Model's Interpretability and Reliability using Biomarkers
Feb 16, 2024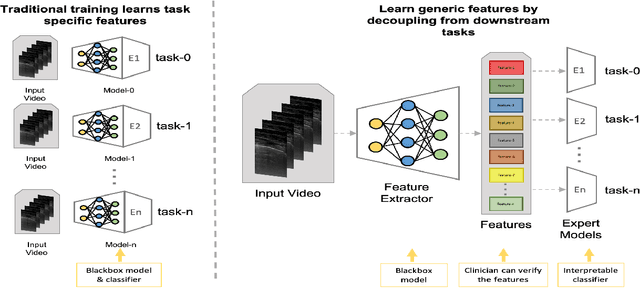

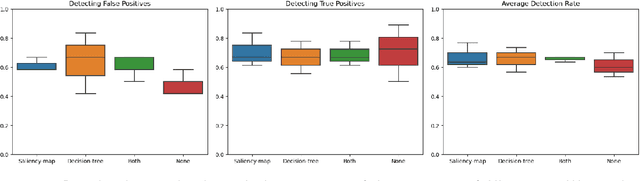

Abstract:Accurate and interpretable diagnostic models are crucial in the safety-critical field of medicine. We investigate the interpretability of our proposed biomarker-based lung ultrasound diagnostic pipeline to enhance clinicians' diagnostic capabilities. The objective of this study is to assess whether explanations from a decision tree classifier, utilizing biomarkers, can improve users' ability to identify inaccurate model predictions compared to conventional saliency maps. Our findings demonstrate that decision tree explanations, based on clinically established biomarkers, can assist clinicians in detecting false positives, thus improving the reliability of diagnostic models in medicine.
Learning Generic Lung Ultrasound Biomarkers for Decoupling Feature Extraction from Downstream Tasks
Jun 16, 2022



Abstract:Contemporary artificial neural networks (ANN) are trained end-to-end, jointly learning both features and classifiers for the task of interest. Though enormously effective, this paradigm imposes significant costs in assembling annotated task-specific datasets and training large-scale networks. We propose to decouple feature learning from downstream lung ultrasound tasks by introducing an auxiliary pre-task of visual biomarker classification. We demonstrate that one can learn an informative, concise, and interpretable feature space from ultrasound videos by training models for predicting biomarker labels. Notably, biomarker feature extractors can be trained from data annotated with weak video-scale supervision. These features can be used by a variety of downstream Expert models targeted for diverse clinical tasks (Diagnosis, lung severity, S/F ratio). Crucially, task-specific expert models are comparable in accuracy to end-to-end models directly trained for such target tasks, while being significantly lower cost to train.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge