Gautam Rajendrakumar Gare
Activation Reward Models for Few-Shot Model Alignment
Jul 02, 2025Abstract:Aligning Large Language Models (LLMs) and Large Multimodal Models (LMMs) to human preferences is a central challenge in improving the quality of the models' generative outputs for real-world applications. A common approach is to use reward modeling to encode preferences, enabling alignment via post-training using reinforcement learning. However, traditional reward modeling is not easily adaptable to new preferences because it requires a separate reward model, commonly trained on large preference datasets. To address this, we introduce Activation Reward Models (Activation RMs) -- a novel few-shot reward modeling method that leverages activation steering to construct well-aligned reward signals using minimal supervision and no additional model finetuning. Activation RMs outperform existing few-shot reward modeling approaches such as LLM-as-a-judge with in-context learning, voting-based scoring, and token probability scoring on standard reward modeling benchmarks. Furthermore, we demonstrate the effectiveness of Activation RMs in mitigating reward hacking behaviors, highlighting their utility for safety-critical applications. Toward this end, we propose PreferenceHack, a novel few-shot setting benchmark, the first to test reward models on reward hacking in a paired preference format. Finally, we show that Activation RM achieves state-of-the-art performance on this benchmark, surpassing even GPT-4o.
Improving Model's Interpretability and Reliability using Biomarkers
Feb 16, 2024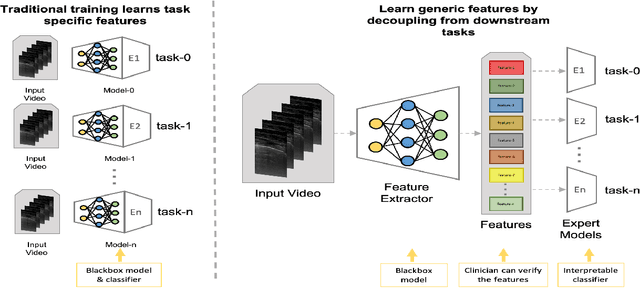

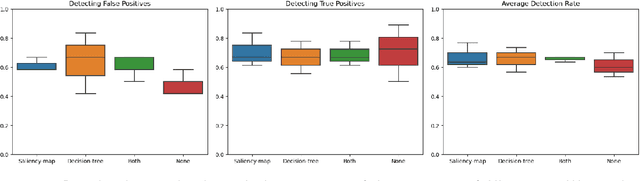

Abstract:Accurate and interpretable diagnostic models are crucial in the safety-critical field of medicine. We investigate the interpretability of our proposed biomarker-based lung ultrasound diagnostic pipeline to enhance clinicians' diagnostic capabilities. The objective of this study is to assess whether explanations from a decision tree classifier, utilizing biomarkers, can improve users' ability to identify inaccurate model predictions compared to conventional saliency maps. Our findings demonstrate that decision tree explanations, based on clinically established biomarkers, can assist clinicians in detecting false positives, thus improving the reliability of diagnostic models in medicine.
Learning Generic Lung Ultrasound Biomarkers for Decoupling Feature Extraction from Downstream Tasks
Jun 16, 2022



Abstract:Contemporary artificial neural networks (ANN) are trained end-to-end, jointly learning both features and classifiers for the task of interest. Though enormously effective, this paradigm imposes significant costs in assembling annotated task-specific datasets and training large-scale networks. We propose to decouple feature learning from downstream lung ultrasound tasks by introducing an auxiliary pre-task of visual biomarker classification. We demonstrate that one can learn an informative, concise, and interpretable feature space from ultrasound videos by training models for predicting biomarker labels. Notably, biomarker feature extractors can be trained from data annotated with weak video-scale supervision. These features can be used by a variety of downstream Expert models targeted for diverse clinical tasks (Diagnosis, lung severity, S/F ratio). Crucially, task-specific expert models are comparable in accuracy to end-to-end models directly trained for such target tasks, while being significantly lower cost to train.
Dense Pixel-Labeling for Reverse-Transfer and Diagnostic Learning on Lung Ultrasound for COVID-19 and Pneumonia Detection
Jan 25, 2022



Abstract:We propose using a pre-trained segmentation model to perform diagnostic classification in order to achieve better generalization and interpretability, terming the technique reverse-transfer learning. We present an architecture to convert segmentation models to classification models. We compare and contrast dense vs sparse segmentation labeling and study its impact on diagnostic classification. We compare the performance of U-Net trained with dense and sparse labels to segment A-lines, B-lines, and Pleural lines on a custom dataset of lung ultrasound scans from 4 patients. Our experiments show that dense labels help reduce false positive detection. We study the classification capability of the dense and sparse trained U-Net and contrast it with a non-pretrained U-Net, to detect and differentiate COVID-19 and Pneumonia on a large ultrasound dataset of about 40k curvilinear and linear probe images. Our segmentation-based models perform better classification when using pretrained segmentation weights, with the dense-label pretrained U-Net performing the best.
* Published in 2021 IEEE 18th International Symposium on Biomedical Imaging (ISBI) \copyright 2021 IEEE
The Role of Pleura and Adipose in Lung Ultrasound AI
Jan 19, 2022Abstract:In this paper, we study the significance of the pleura and adipose tissue in lung ultrasound AI analysis. We highlight their more prominent appearance when using high-frequency linear (HFL) instead of curvilinear ultrasound probes, showing HFL reveals better pleura detail. We compare the diagnostic utility of the pleura and adipose tissue using an HFL ultrasound probe. Masking the adipose tissue during training and inference (while retaining the pleural line and Merlin's space artifacts such as A-lines and B-lines) improved the AI model's diagnostic accuracy.
* Published in MICCAI 2021 workshop on Lessons Learned from the development and application of medical imaging-based AI technologies for combating COVID-19 (LL-COVID19). The first two authors contributed equally to this work
Weakly Supervised Contrastive Learning for Better Severity Scoring of Lung Ultrasound
Jan 18, 2022



Abstract:With the onset of the COVID-19 pandemic, ultrasound has emerged as an effective tool for bedside monitoring of patients. Due to this, a large amount of lung ultrasound scans have been made available which can be used for AI based diagnosis and analysis. Several AI-based patient severity scoring models have been proposed that rely on scoring the appearance of the ultrasound scans. AI models are trained using ultrasound-appearance severity scores that are manually labeled based on standardized visual features. We address the challenge of labeling every ultrasound frame in the video clips. Our contrastive learning method treats the video clip severity labels as noisy weak severity labels for individual frames, thus requiring only video-level labels. We show that it performs better than the conventional cross-entropy loss based training. We combine frame severity predictions to come up with video severity predictions and show that the frame based model achieves comparable performance to a video based TSM model, on a large dataset combining public and private sources.
Exploiting Class Similarity for Machine Learning with Confidence Labels and Projective Loss Functions
Mar 25, 2021



Abstract:Class labels used for machine learning are relatable to each other, with certain class labels being more similar to each other than others (e.g. images of cats and dogs are more similar to each other than those of cats and cars). Such similarity among classes is often the cause of poor model performance due to the models confusing between them. Current labeling techniques fail to explicitly capture such similarity information. In this paper, we instead exploit the similarity between classes by capturing the similarity information with our novel confidence labels. Confidence labels are probabilistic labels denoting the likelihood of similarity, or confusability, between the classes. Often even after models are trained to differentiate between classes in the feature space, the similar classes' latent space still remains clustered. We view this type of clustering as valuable information and exploit it with our novel projective loss functions. Our projective loss functions are designed to work with confidence labels with an ability to relax the loss penalty for errors that confuse similar classes. We use our approach to train neural networks with noisy labels, as we believe noisy labels are partly a result of confusability arising from class similarity. We show improved performance compared to the use of standard loss functions. We conduct a detailed analysis using the CIFAR-10 dataset and show our proposed methods' applicability to larger datasets, such as ImageNet and Food-101N.
W-Net: Dense Semantic Segmentation of Subcutaneous Tissue in Ultrasound Images by Expanding U-Net to Incorporate Ultrasound RF Waveform Data
Sep 02, 2020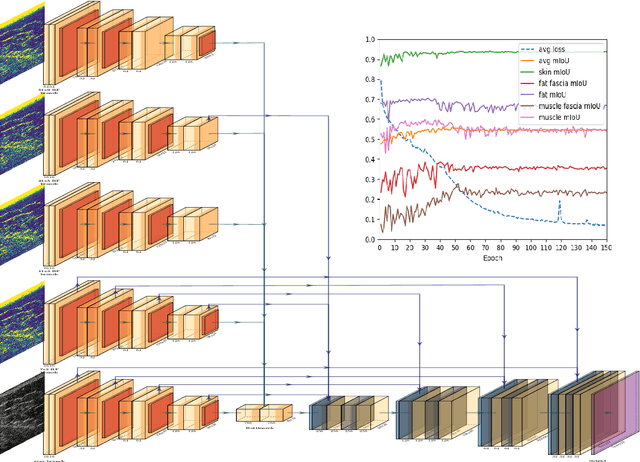
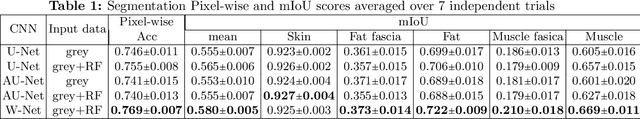
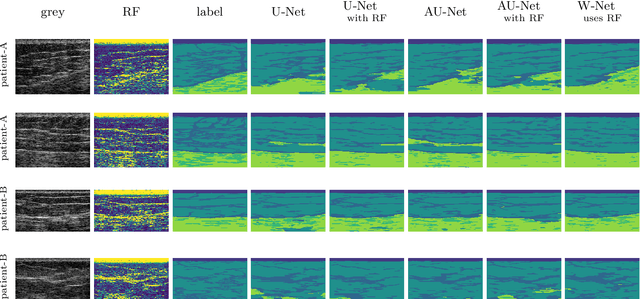
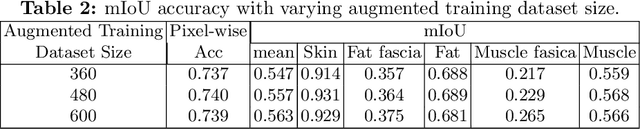
Abstract:We present W-Net, a novel Convolution Neural Network (CNN) framework that employs raw ultrasound waveforms from each A-scan, typically referred to as ultrasound Radio Frequency (RF) data, in addition to the gray ultrasound image to semantically segment and label tissues. Unlike prior work, we seek to label every pixel in the image, without the use of a background class. To the best of our knowledge, this is also the first deep-learning or CNN approach for segmentation that analyses ultrasound raw RF data along with the gray image. International patent(s) pending [PCT/US20/37519]. We chose subcutaneous tissue (SubQ) segmentation as our initial clinical goal since it has diverse intermixed tissues, is challenging to segment, and is an underrepresented research area. SubQ potential applications include plastic surgery, adipose stem-cell harvesting, lymphatic monitoring, and possibly detection/treatment of certain types of tumors. A custom dataset consisting of hand-labeled images by an expert clinician and trainees are used for the experimentation, currently labeled into the following categories: skin, fat, fat fascia/stroma, muscle and muscle fascia. We compared our results with U-Net and Attention U-Net. Our novel \emph{W-Net}'s RF-Waveform input and architecture increased mIoU accuracy (averaged across all tissue classes) by 4.5\% and 4.9\% compared to regular U-Net and Attention U-Net, respectively. We present analysis as to why the Muscle fascia and Fat fascia/stroma are the most difficult tissues to label. Muscle fascia in particular, the most difficult anatomic class to recognize for both humans and AI algorithms, saw mIoU improvements of 13\% and 16\% from our W-Net vs U-Net and Attention U-Net respectively.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge