Kamalaker Dadi
PARIETAL
HyperGALE: ASD Classification via Hypergraph Gated Attention with Learnable Hyperedges
Mar 21, 2024Abstract:Autism Spectrum Disorder (ASD) is a neurodevelopmental condition characterized by varied social cognitive challenges and repetitive behavioral patterns. Identifying reliable brain imaging-based biomarkers for ASD has been a persistent challenge due to the spectrum's diverse symptomatology. Existing baselines in the field have made significant strides in this direction, yet there remains room for improvement in both performance and interpretability. We propose \emph{HyperGALE}, which builds upon the hypergraph by incorporating learned hyperedges and gated attention mechanisms. This approach has led to substantial improvements in the model's ability to interpret complex brain graph data, offering deeper insights into ASD biomarker characterization. Evaluated on the extensive ABIDE II dataset, \emph{HyperGALE} not only improves interpretability but also demonstrates statistically significant enhancements in key performance metrics compared to both previous baselines and the foundational hypergraph model. The advancement \emph{HyperGALE} brings to ASD research highlights the potential of sophisticated graph-based techniques in neurodevelopmental studies. The source code and implementation instructions are available at GitHub:https://github.com/mehular0ra/HyperGALE.
BeSt-LeS: Benchmarking Stroke Lesion Segmentation using Deep Supervision
Oct 10, 2023Abstract:Brain stroke has become a significant burden on global health and thus we need remedies and prevention strategies to overcome this challenge. For this, the immediate identification of stroke and risk stratification is the primary task for clinicians. To aid expert clinicians, automated segmentation models are crucial. In this work, we consider the publicly available dataset ATLAS $v2.0$ to benchmark various end-to-end supervised U-Net style models. Specifically, we have benchmarked models on both 2D and 3D brain images and evaluated them using standard metrics. We have achieved the highest Dice score of 0.583 on the 2D transformer-based model and 0.504 on the 3D residual U-Net respectively. We have conducted the Wilcoxon test for 3D models to correlate the relationship between predicted and actual stroke volume. For reproducibility, the code and model weights are made publicly available: https://github.com/prantik-pdeb/BeSt-LeS.
Fine-grain atlases of functional modes for fMRI analysis
Mar 05, 2020
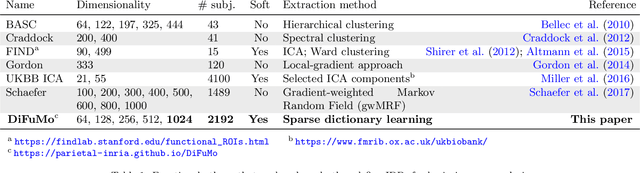
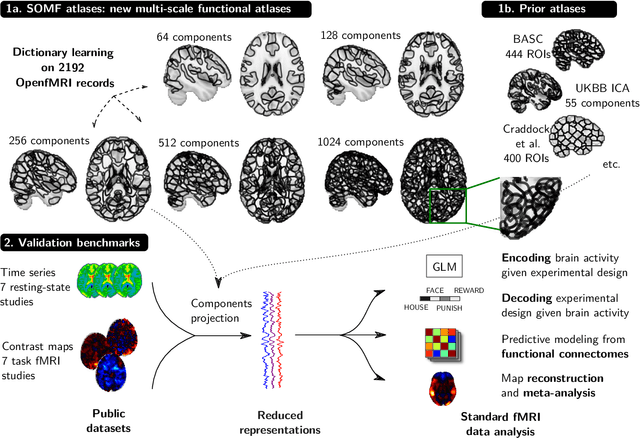
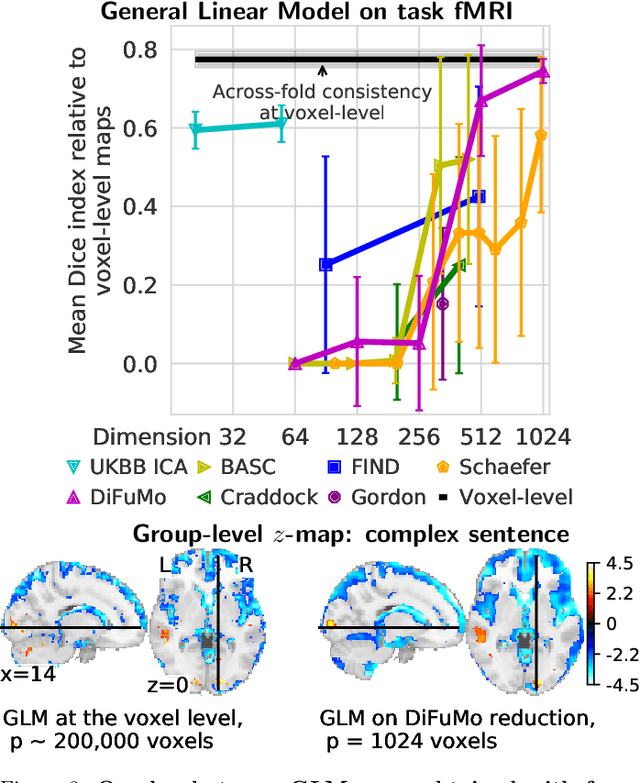
Abstract:Population imaging markedly increased the size of functional-imaging datasets, shedding new light on the neural basis of inter-individual differences. Analyzing these large data entails new scalability challenges, computational and statistical. For this reason, brain images are typically summarized in a few signals, for instance reducing voxel-level measures with brain atlases or functional modes. A good choice of the corresponding brain networks is important, as most data analyses start from these reduced signals. We contribute finely-resolved atlases of functional modes, comprising from 64 to 1024 networks. These dictionaries of functional modes (DiFuMo) are trained on millions of fMRI functional brain volumes of total size 2.4TB, spanned over 27 studies and many research groups. We demonstrate the benefits of extracting reduced signals on our fine-grain atlases for many classic functional data analysis pipelines: stimuli decoding from 12,334 brain responses, standard GLM analysis of fMRI across sessions and individuals, extraction of resting-state functional-connectomes biomarkers for 2,500 individuals, data compression and meta-analysis over more than 15,000 statistical maps. In each of these analysis scenarii, we compare the performance of our functional atlases with that of other popular references, and to a simple voxel-level analysis. Results highlight the importance of using high-dimensional "soft" functional atlases, to represent and analyse brain activity while capturing its functional gradients. Analyses on high-dimensional modes achieve similar statistical performance as at the voxel level, but with much reduced computational cost and higher interpretability. In addition to making them available, we provide meaningful names for these modes, based on their anatomical location. It will facilitate reporting of results.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge