Krzysztof J. Gorgolewski
PARIETAL
DeepDefacer: Automatic Removal of Facial Features via U-Net Image Segmentation
May 31, 2022



Abstract:Recent advancements in the field of magnetic resonance imaging (MRI) have enabled large-scale collaboration among clinicians and researchers for neuroimaging tasks. However, researchers are often forced to use outdated and slow software to anonymize MRI images for publication. These programs specifically perform expensive mathematical operations over 3D images that rapidly slow down anonymization speed as an image's volume increases in size. In this paper, we introduce DeepDefacer, an application of deep learning to MRI anonymization that uses a streamlined 3D U-Net network to mask facial regions in MRI images with a significant increase in speed over traditional de-identification software. We train DeepDefacer on MRI images from the Brain Development Organization (IXI) and International Consortium for Brain Mapping (ICBM) and quantitatively evaluate our model against a baseline 3D U-Net model with regards to Dice, recall, and precision scores. We also evaluate DeepDefacer against Pydeface, a traditional defacing application, with regards to speed on a range of CPU and GPU devices and qualitatively evaluate our model's defaced output versus the ground truth images produced by Pydeface. We provide a link to a PyPi program at the end of this manuscript to encourage further research into the application of deep learning to MRI anonymization.
Fine-grain atlases of functional modes for fMRI analysis
Mar 05, 2020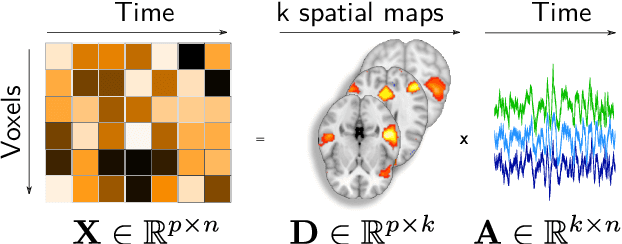
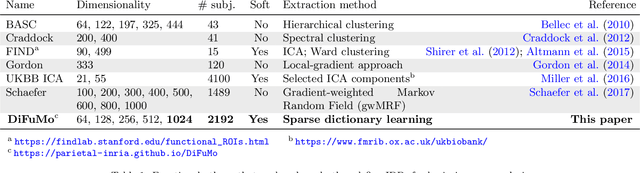
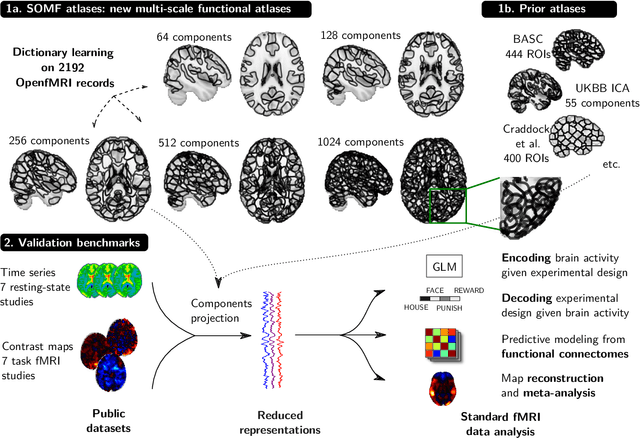
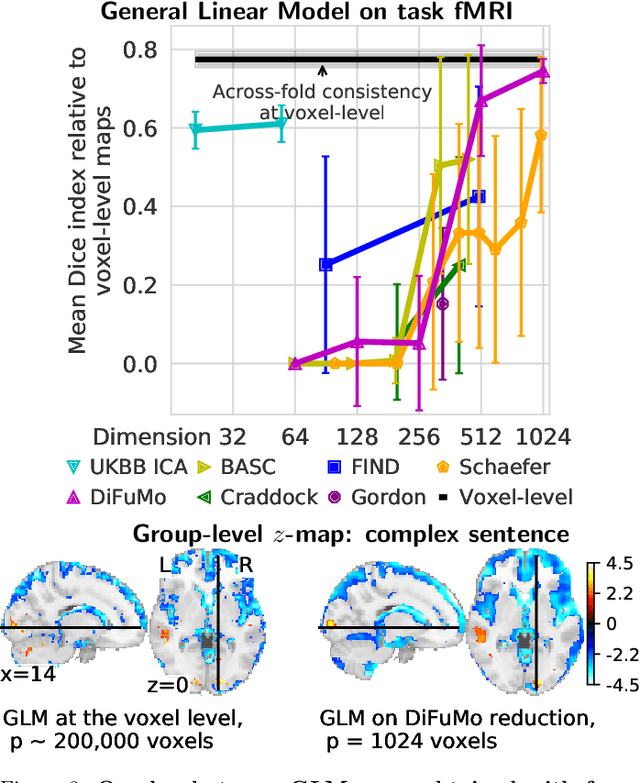
Abstract:Population imaging markedly increased the size of functional-imaging datasets, shedding new light on the neural basis of inter-individual differences. Analyzing these large data entails new scalability challenges, computational and statistical. For this reason, brain images are typically summarized in a few signals, for instance reducing voxel-level measures with brain atlases or functional modes. A good choice of the corresponding brain networks is important, as most data analyses start from these reduced signals. We contribute finely-resolved atlases of functional modes, comprising from 64 to 1024 networks. These dictionaries of functional modes (DiFuMo) are trained on millions of fMRI functional brain volumes of total size 2.4TB, spanned over 27 studies and many research groups. We demonstrate the benefits of extracting reduced signals on our fine-grain atlases for many classic functional data analysis pipelines: stimuli decoding from 12,334 brain responses, standard GLM analysis of fMRI across sessions and individuals, extraction of resting-state functional-connectomes biomarkers for 2,500 individuals, data compression and meta-analysis over more than 15,000 statistical maps. In each of these analysis scenarii, we compare the performance of our functional atlases with that of other popular references, and to a simple voxel-level analysis. Results highlight the importance of using high-dimensional "soft" functional atlases, to represent and analyse brain activity while capturing its functional gradients. Analyses on high-dimensional modes achieve similar statistical performance as at the voxel level, but with much reduced computational cost and higher interpretability. In addition to making them available, we provide meaningful names for these modes, based on their anatomical location. It will facilitate reporting of results.
Computational and informatics advances for reproducible data analysis in neuroimaging
Sep 24, 2018



Abstract:The reproducibility of scientific research has become a point of critical concern. We argue that openness and transparency are critical for reproducibility, and we outline an ecosystem for open and transparent science that has emerged within the human neuroimaging community. We discuss the range of open data sharing resources that have been developed for neuroimaging data, and the role of data standards (particularly the Brain Imaging Data Structure) in enabling the automated sharing, processing, and reuse of large neuroimaging datasets. We outline how the open-source Python language has provided the basis for a data science platform that enables reproducible data analysis and visualization. We also discuss how new advances in software engineering, such as containerization, provide the basis for greater reproducibility in data analysis. The emergence of this new ecosystem provides an example for many areas of science that are currently struggling with reproducibility.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge