Marco Guevara
Improving Clinical NLP Performance through Language Model-Generated Synthetic Clinical Data
Mar 28, 2024
Abstract:Generative models have been showing potential for producing data in mass. This study explores the enhancement of clinical natural language processing performance by utilizing synthetic data generated from advanced language models. Promising results show feasible applications in such a high-stakes domain.
The impact of using an AI chatbot to respond to patient messages
Oct 26, 2023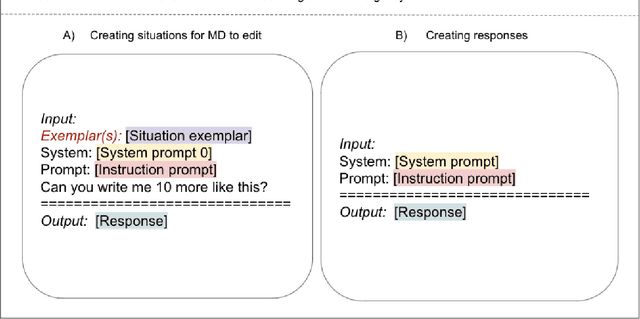
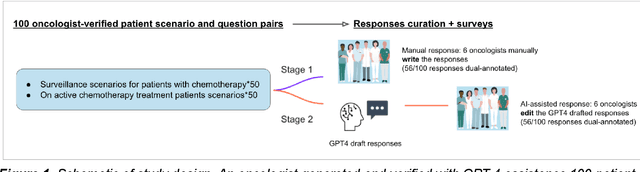
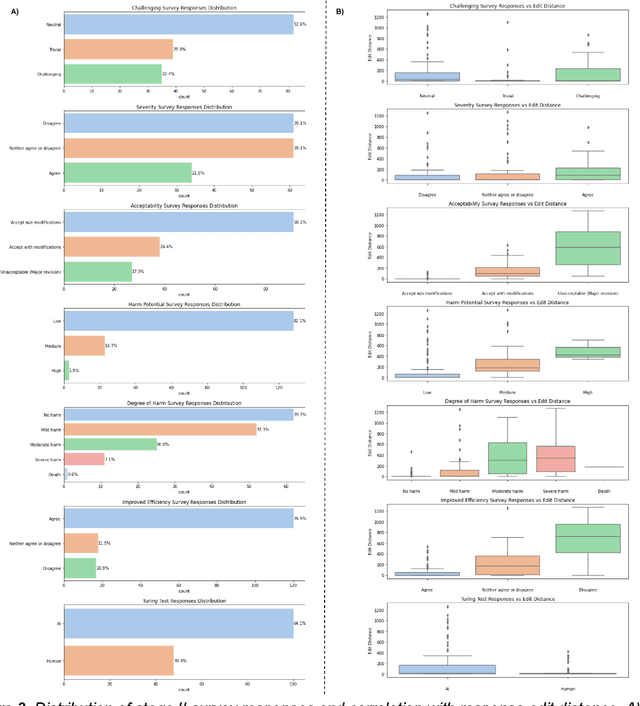
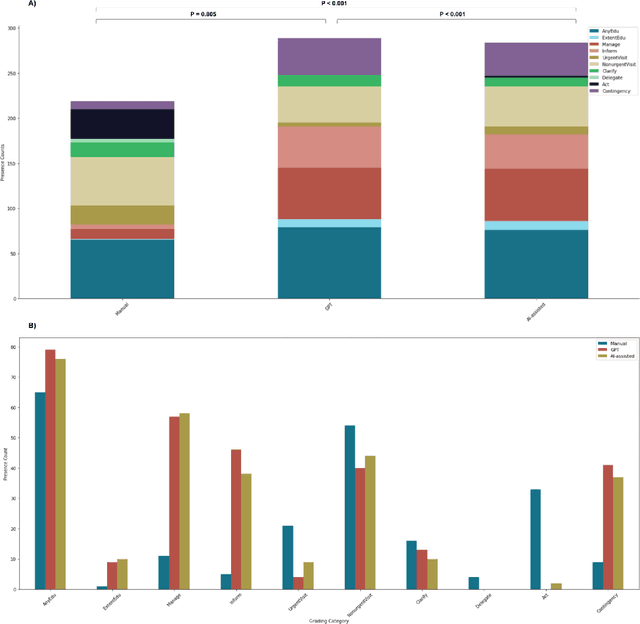
Abstract:Documentation burden is a major contributor to clinician burnout, which is rising nationally and is an urgent threat to our ability to care for patients. Artificial intelligence (AI) chatbots, such as ChatGPT, could reduce clinician burden by assisting with documentation. Although many hospitals are actively integrating such systems into electronic medical record systems, AI chatbots utility and impact on clinical decision-making have not been studied for this intended use. We are the first to examine the utility of large language models in assisting clinicians draft responses to patient questions. In our two-stage cross-sectional study, 6 oncologists responded to 100 realistic synthetic cancer patient scenarios and portal messages developed to reflect common medical situations, first manually, then with AI assistance. We find AI-assisted responses were longer, less readable, but provided acceptable drafts without edits 58% of time. AI assistance improved efficiency 77% of time, with low harm risk (82% safe). However, 7.7% unedited AI responses could severely harm. In 31% cases, physicians thought AI drafts were human-written. AI assistance led to more patient education recommendations, fewer clinical actions than manual responses. Results show promise for AI to improve clinician efficiency and patient care through assisting documentation, if used judiciously. Monitoring model outputs and human-AI interaction remains crucial for safe implementation.
Large Language Models to Identify Social Determinants of Health in Electronic Health Records
Aug 11, 2023Abstract:Social determinants of health (SDoH) have an important impact on patient outcomes but are incompletely collected from the electronic health records (EHR). This study researched the ability of large language models to extract SDoH from free text in EHRs, where they are most commonly documented, and explored the role of synthetic clinical text for improving the extraction of these scarcely documented, yet extremely valuable, clinical data. 800 patient notes were annotated for SDoH categories, and several transformer-based models were evaluated. The study also experimented with synthetic data generation and assessed for algorithmic bias. Our best-performing models were fine-tuned Flan-T5 XL (macro-F1 0.71) for any SDoH, and Flan-T5 XXL (macro-F1 0.70). The benefit of augmenting fine-tuning with synthetic data varied across model architecture and size, with smaller Flan-T5 models (base and large) showing the greatest improvements in performance (delta F1 +0.12 to +0.23). Model performance was similar on the in-hospital system dataset but worse on the MIMIC-III dataset. Our best-performing fine-tuned models outperformed zero- and few-shot performance of ChatGPT-family models for both tasks. These fine-tuned models were less likely than ChatGPT to change their prediction when race/ethnicity and gender descriptors were added to the text, suggesting less algorithmic bias (p<0.05). At the patient-level, our models identified 93.8% of patients with adverse SDoH, while ICD-10 codes captured 2.0%. Our method can effectively extracted SDoH information from clinic notes, performing better compare to GPT zero- and few-shot settings. These models could enhance real-world evidence on SDoH and aid in identifying patients needing social support.
Natural language processing to automatically extract the presence and severity of esophagitis in notes of patients undergoing radiotherapy
Mar 24, 2023Abstract:Radiotherapy (RT) toxicities can impair survival and quality-of-life, yet remain under-studied. Real-world evidence holds potential to improve our understanding of toxicities, but toxicity information is often only in clinical notes. We developed natural language processing (NLP) models to identify the presence and severity of esophagitis from notes of patients treated with thoracic RT. We fine-tuned statistical and pre-trained BERT-based models for three esophagitis classification tasks: Task 1) presence of esophagitis, Task 2) severe esophagitis or not, and Task 3) no esophagitis vs. grade 1 vs. grade 2-3. Transferability was tested on 345 notes from patients with esophageal cancer undergoing RT. Fine-tuning PubmedBERT yielded the best performance. The best macro-F1 was 0.92, 0.82, and 0.74 for Task 1, 2, and 3, respectively. Selecting the most informative note sections during fine-tuning improved macro-F1 by over 2% for all tasks. Silver-labeled data improved the macro-F1 by over 3% across all tasks. For the esophageal cancer notes, the best macro-F1 was 0.73, 0.74, and 0.65 for Task 1, 2, and 3, respectively, without additional fine-tuning. To our knowledge, this is the first effort to automatically extract esophagitis toxicity severity according to CTCAE guidelines from clinic notes. The promising performance provides proof-of-concept for NLP-based automated detailed toxicity monitoring in expanded domains.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge