Yanjun Gao
Kenneth
Race, Ethnicity and Their Implication on Bias in Large Language Models
Jan 19, 2026Abstract:Large language models (LLMs) increasingly operate in high-stakes settings including healthcare and medicine, where demographic attributes such as race and ethnicity may be explicitly stated or implicitly inferred from text. However, existing studies primarily document outcome-level disparities, offering limited insight into internal mechanisms underlying these effects. We present a mechanistic study of how race and ethnicity are represented and operationalized within LLMs. Using two publicly available datasets spanning toxicity-related generation and clinical narrative understanding tasks, we analyze three open-source models with a reproducible interpretability pipeline combining probing, neuron-level attribution, and targeted intervention. We find that demographic information is distributed across internal units with substantial cross-model variation. Although some units encode sensitive or stereotype-related associations from pretraining, identical demographic cues can induce qualitatively different behaviors. Interventions suppressing such neurons reduce bias but leave substantial residual effects, suggesting behavioral rather than representational change and motivating more systematic mitigation.
Evaluating Retrieval-Augmented Generation vs. Long-Context Input for Clinical Reasoning over EHRs
Aug 20, 2025Abstract:Electronic health records (EHRs) are long, noisy, and often redundant, posing a major challenge for the clinicians who must navigate them. Large language models (LLMs) offer a promising solution for extracting and reasoning over this unstructured text, but the length of clinical notes often exceeds even state-of-the-art models' extended context windows. Retrieval-augmented generation (RAG) offers an alternative by retrieving task-relevant passages from across the entire EHR, potentially reducing the amount of required input tokens. In this work, we propose three clinical tasks designed to be replicable across health systems with minimal effort: 1) extracting imaging procedures, 2) generating timelines of antibiotic use, and 3) identifying key diagnoses. Using EHRs from actual hospitalized patients, we test three state-of-the-art LLMs with varying amounts of provided context, using either targeted text retrieval or the most recent clinical notes. We find that RAG closely matches or exceeds the performance of using recent notes, and approaches the performance of using the models' full context while requiring drastically fewer input tokens. Our results suggest that RAG remains a competitive and efficient approach even as newer models become capable of handling increasingly longer amounts of text.
An Information-Theoretic Perspective on Multi-LLM Uncertainty Estimation
Jul 09, 2025Abstract:Large language models (LLMs) often behave inconsistently across inputs, indicating uncertainty and motivating the need for its quantification in high-stakes settings. Prior work on calibration and uncertainty quantification often focuses on individual models, overlooking the potential of model diversity. We hypothesize that LLMs make complementary predictions due to differences in training and the Zipfian nature of language, and that aggregating their outputs leads to more reliable uncertainty estimates. To leverage this, we propose MUSE (Multi-LLM Uncertainty via Subset Ensembles), a simple information-theoretic method that uses Jensen-Shannon Divergence to identify and aggregate well-calibrated subsets of LLMs. Experiments on binary prediction tasks demonstrate improved calibration and predictive performance compared to single-model and naive ensemble baselines.
Uncovering Hidden Violent Tendencies in LLMs: A Demographic Analysis via Behavioral Vignettes
Jun 25, 2025Abstract:Large language models (LLMs) are increasingly proposed for detecting and responding to violent content online, yet their ability to reason about morally ambiguous, real-world scenarios remains underexamined. We present the first study to evaluate LLMs using a validated social science instrument designed to measure human response to everyday conflict, namely the Violent Behavior Vignette Questionnaire (VBVQ). To assess potential bias, we introduce persona-based prompting that varies race, age, and geographic identity within the United States. Six LLMs developed across different geopolitical and organizational contexts are evaluated under a unified zero-shot setting. Our study reveals two key findings: (1) LLMs surface-level text generation often diverges from their internal preference for violent responses; (2) their violent tendencies vary across demographics, frequently contradicting established findings in criminology, social science, and psychology.
Attributing Response to Context: A Jensen-Shannon Divergence Driven Mechanistic Study of Context Attribution in Retrieval-Augmented Generation
May 22, 2025Abstract:Retrieval-Augmented Generation (RAG) leverages large language models (LLMs) combined with external contexts to enhance the accuracy and reliability of generated responses. However, reliably attributing generated content to specific context segments, context attribution, remains challenging due to the computationally intensive nature of current methods, which often require extensive fine-tuning or human annotation. In this work, we introduce a novel Jensen-Shannon Divergence driven method to Attribute Response to Context (ARC-JSD), enabling efficient and accurate identification of essential context sentences without additional fine-tuning or surrogate modelling. Evaluations on a wide range of RAG benchmarks, such as TyDi QA, Hotpot QA, and Musique, using instruction-tuned LLMs in different scales demonstrate superior accuracy and significant computational efficiency improvements compared to the previous surrogate-based method. Furthermore, our mechanistic analysis reveals specific attention heads and multilayer perceptron (MLP) layers responsible for context attribution, providing valuable insights into the internal workings of RAG models.
"It Felt Like I Was Left in the Dark": Exploring Information Needs and Design Opportunities for Family Caregivers of Older Adult Patients in Critical Care Settings
Feb 07, 2025Abstract:Older adult patients constitute a rapidly growing subgroup of Intensive Care Unit (ICU) patients. In these situations, their family caregivers are expected to represent the unconscious patients to access and interpret patients' medical information. However, caregivers currently have to rely on overloaded clinicians for information updates and typically lack the health literacy to understand complex medical information. Our project aims to explore the information needs of caregivers of ICU older adult patients, from which we can propose design opportunities to guide future AI systems. The project begins with formative interviews with 11 caregivers to identify their challenges in accessing and interpreting medical information; From these findings, we then synthesize design requirements and propose an AI system prototype to cope with caregivers' challenges. The system prototype has two key features: a timeline visualization to show the AI extracted and summarized older adult patients' key medical events; and an LLM-based chatbot to provide context-aware informational support. We conclude our paper by reporting on the follow-up user evaluation of the system and discussing future AI-based systems for ICU caregivers of older adults.
Zero-shot Large Language Models for Long Clinical Text Summarization with Temporal Reasoning
Jan 30, 2025



Abstract:Recent advancements in large language models (LLMs) have shown potential for transforming data processing in healthcare, particularly in understanding complex clinical narratives. This study evaluates the efficacy of zero-shot LLMs in summarizing long clinical texts that require temporal reasoning, a critical aspect for comprehensively capturing patient histories and treatment trajectories. We applied a series of advanced zero-shot LLMs to extensive clinical documents, assessing their ability to integrate and accurately reflect temporal dynamics without prior task-specific training. While the models efficiently identified key temporal events, they struggled with chronological coherence over prolonged narratives. The evaluation, combining quantitative and qualitative methods, highlights the strengths and limitations of zero-shot LLMs in clinical text summarization. The results suggest that while promising, zero-shot LLMs require further refinement to effectively support clinical decision-making processes, underscoring the need for enhanced model training approaches that better capture the nuances of temporal information in long context medical documents.
Development and Validation of the Provider Documentation Summarization Quality Instrument for Large Language Models
Jan 15, 2025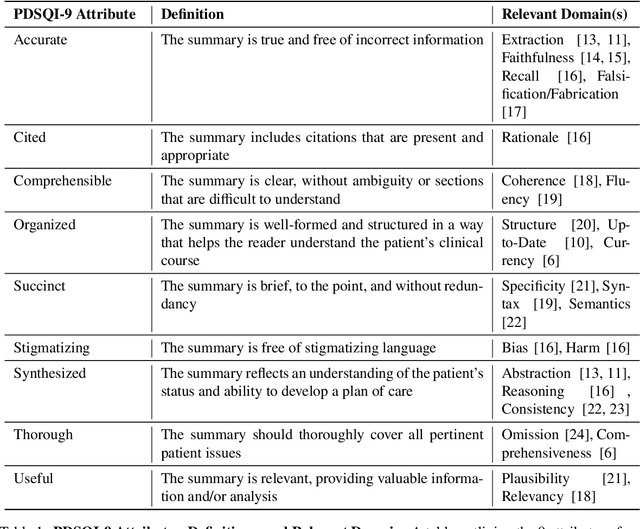
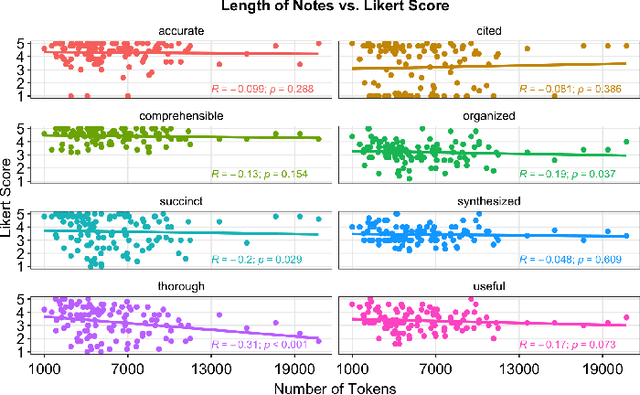
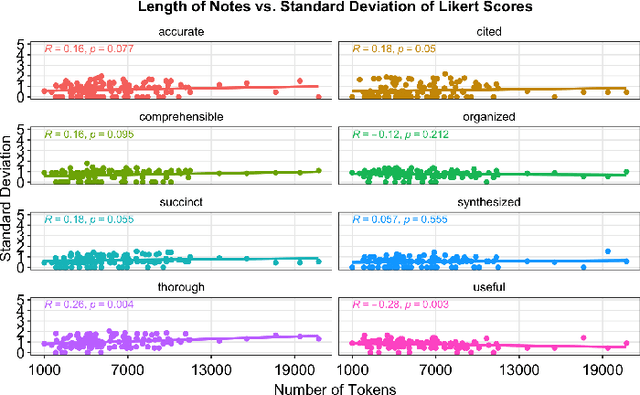
Abstract:As Large Language Models (LLMs) are integrated into electronic health record (EHR) workflows, validated instruments are essential to evaluate their performance before implementation. Existing instruments for provider documentation quality are often unsuitable for the complexities of LLM-generated text and lack validation on real-world data. The Provider Documentation Summarization Quality Instrument (PDSQI-9) was developed to evaluate LLM-generated clinical summaries. Multi-document summaries were generated from real-world EHR data across multiple specialties using several LLMs (GPT-4o, Mixtral 8x7b, and Llama 3-8b). Validation included Pearson correlation for substantive validity, factor analysis and Cronbach's alpha for structural validity, inter-rater reliability (ICC and Krippendorff's alpha) for generalizability, a semi-Delphi process for content validity, and comparisons of high- versus low-quality summaries for discriminant validity. Seven physician raters evaluated 779 summaries and answered 8,329 questions, achieving over 80% power for inter-rater reliability. The PDSQI-9 demonstrated strong internal consistency (Cronbach's alpha = 0.879; 95% CI: 0.867-0.891) and high inter-rater reliability (ICC = 0.867; 95% CI: 0.867-0.868), supporting structural validity and generalizability. Factor analysis identified a 4-factor model explaining 58% of the variance, representing organization, clarity, accuracy, and utility. Substantive validity was supported by correlations between note length and scores for Succinct (rho = -0.200, p = 0.029) and Organized (rho = -0.190, p = 0.037). Discriminant validity distinguished high- from low-quality summaries (p < 0.001). The PDSQI-9 demonstrates robust construct validity, supporting its use in clinical practice to evaluate LLM-generated summaries and facilitate safer integration of LLMs into healthcare workflows.
Position Paper On Diagnostic Uncertainty Estimation from Large Language Models: Next-Word Probability Is Not Pre-test Probability
Nov 07, 2024Abstract:Large language models (LLMs) are being explored for diagnostic decision support, yet their ability to estimate pre-test probabilities, vital for clinical decision-making, remains limited. This study evaluates two LLMs, Mistral-7B and Llama3-70B, using structured electronic health record data on three diagnosis tasks. We examined three current methods of extracting LLM probability estimations and revealed their limitations. We aim to highlight the need for improved techniques in LLM confidence estimation.
When Raw Data Prevails: Are Large Language Model Embeddings Effective in Numerical Data Representation for Medical Machine Learning Applications?
Aug 15, 2024



Abstract:The introduction of Large Language Models (LLMs) has advanced data representation and analysis, bringing significant progress in their use for medical questions and answering. Despite these advancements, integrating tabular data, especially numerical data pivotal in clinical contexts, into LLM paradigms has not been thoroughly explored. In this study, we examine the effectiveness of vector representations from last hidden states of LLMs for medical diagnostics and prognostics using electronic health record (EHR) data. We compare the performance of these embeddings with that of raw numerical EHR data when used as feature inputs to traditional machine learning (ML) algorithms that excel at tabular data learning, such as eXtreme Gradient Boosting. We focus on instruction-tuned LLMs in a zero-shot setting to represent abnormal physiological data and evaluating their utilities as feature extractors to enhance ML classifiers for predicting diagnoses, length of stay, and mortality. Furthermore, we examine prompt engineering techniques on zero-shot and few-shot LLM embeddings to measure their impact comprehensively. Although findings suggest the raw data features still prevails in medical ML tasks, zero-shot LLM embeddings demonstrate competitive results, suggesting a promising avenue for future research in medical applications.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge