Timothy A Miller
Position Paper On Diagnostic Uncertainty Estimation from Large Language Models: Next-Word Probability Is Not Pre-test Probability
Nov 07, 2024Abstract:Large language models (LLMs) are being explored for diagnostic decision support, yet their ability to estimate pre-test probabilities, vital for clinical decision-making, remains limited. This study evaluates two LLMs, Mistral-7B and Llama3-70B, using structured electronic health record data on three diagnosis tasks. We examined three current methods of extracting LLM probability estimations and revealed their limitations. We aim to highlight the need for improved techniques in LLM confidence estimation.
When Raw Data Prevails: Are Large Language Model Embeddings Effective in Numerical Data Representation for Medical Machine Learning Applications?
Aug 15, 2024



Abstract:The introduction of Large Language Models (LLMs) has advanced data representation and analysis, bringing significant progress in their use for medical questions and answering. Despite these advancements, integrating tabular data, especially numerical data pivotal in clinical contexts, into LLM paradigms has not been thoroughly explored. In this study, we examine the effectiveness of vector representations from last hidden states of LLMs for medical diagnostics and prognostics using electronic health record (EHR) data. We compare the performance of these embeddings with that of raw numerical EHR data when used as feature inputs to traditional machine learning (ML) algorithms that excel at tabular data learning, such as eXtreme Gradient Boosting. We focus on instruction-tuned LLMs in a zero-shot setting to represent abnormal physiological data and evaluating their utilities as feature extractors to enhance ML classifiers for predicting diagnoses, length of stay, and mortality. Furthermore, we examine prompt engineering techniques on zero-shot and few-shot LLM embeddings to measure their impact comprehensively. Although findings suggest the raw data features still prevails in medical ML tasks, zero-shot LLM embeddings demonstrate competitive results, suggesting a promising avenue for future research in medical applications.
Confederated Machine Learning on Horizontally and Vertically Separated Medical Data for Large-Scale Health System Intelligence
Oct 04, 2019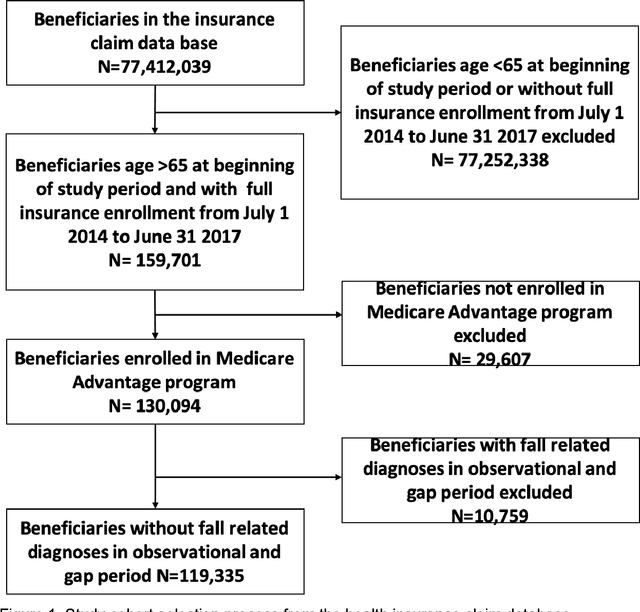
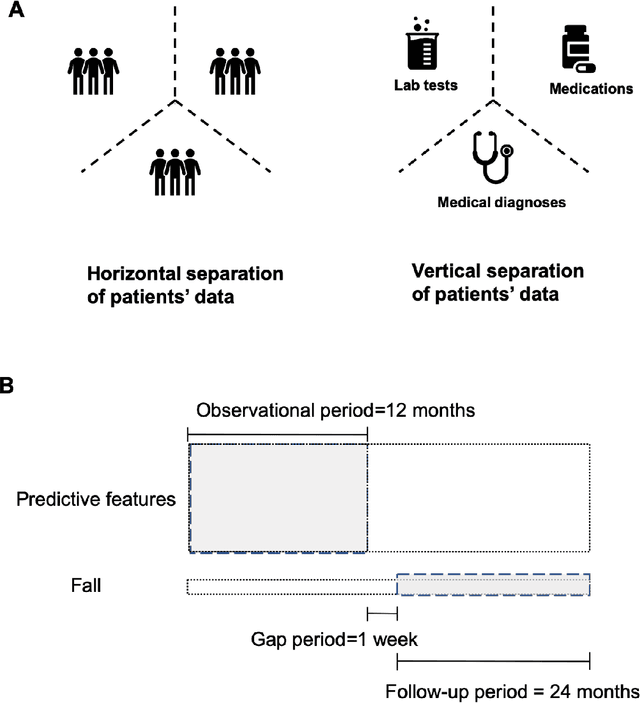
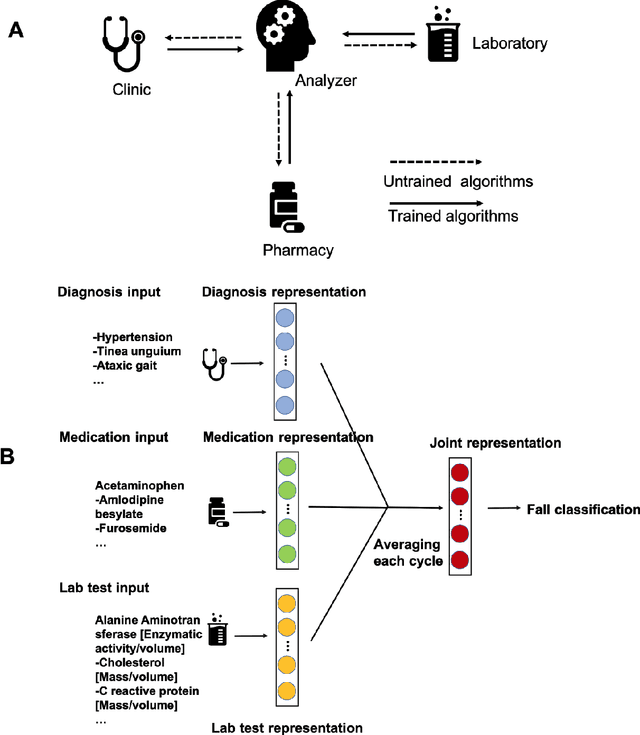
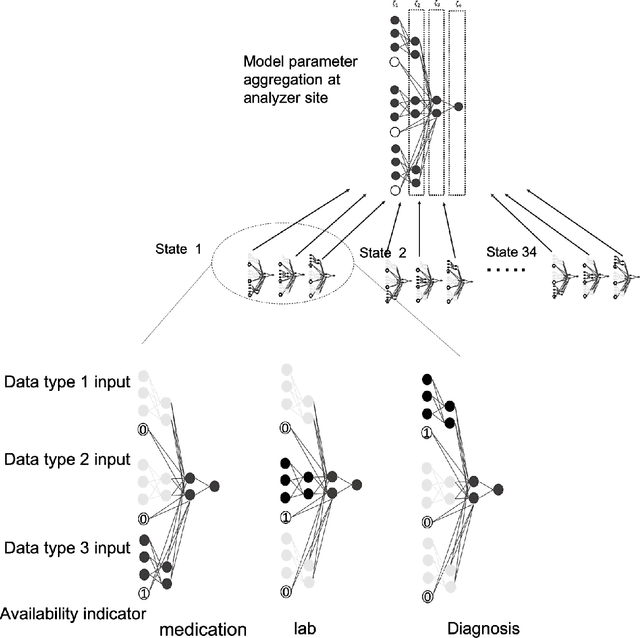
Abstract:A patient's health information is generally fragmented across silos. Though it is technically feasible to unite data for analysis in a manner that underpins a rapid learning healthcare system, privacy concerns and regulatory barriers limit data centralization. Machine learning can be conducted in a federated manner on patient datasets with the same set of variables, but separated across sites of care. But federated learning cannot handle the situation where different data types for a given patient are separated vertically across different organizations. We call methods that enable machine learning model training on data separated by two or more degrees "confederated machine learning." We built and evaluated a confederated machine learning model to stratify the risk of accidental falls among the elderly
Simplified Neural Unsupervised Domain Adaptation
May 22, 2019


Abstract:Unsupervised domain adaptation (UDA) is the task of modifying a statistical model trained on labeled data from a source domain to achieve better performance on data from a target domain, with access to only unlabeled data in the target domain. Existing state-of-the-art UDA approaches use neural networks to learn representations that can predict the values of subset of important features called "pivot features." In this work, we show that it is possible to improve on these methods by jointly training the representation learner with the task learner, and examine the importance of existing pivot selection methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge