Kumaradevan Punithakumar
Accelerated 3D-3D rigid registration of echocardiographic images obtained from apical window using particle filter
Apr 28, 2025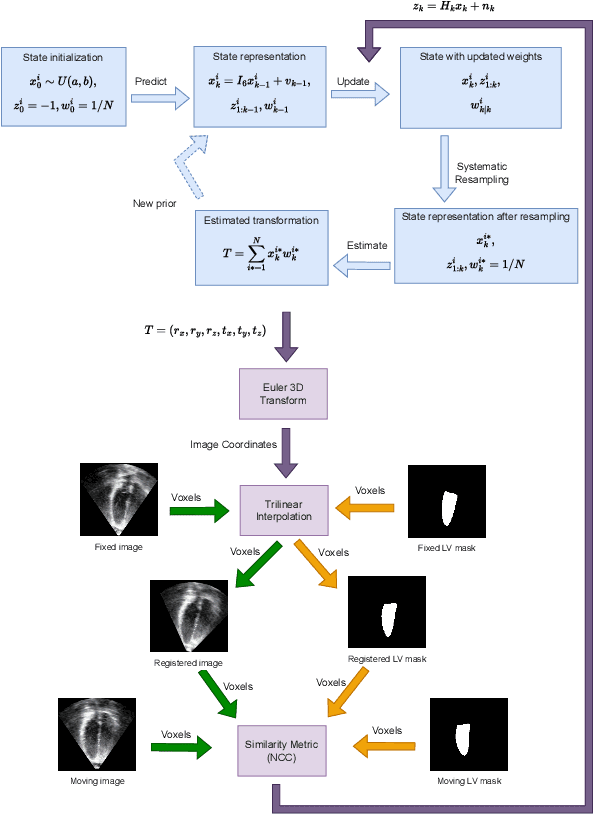



Abstract:The perfect alignment of 3D echocardiographic images captured from various angles has improved image quality and broadened the field of view. This study proposes an accelerated sequential Monte Carlo (SMC) algorithm for 3D-3D rigid registration of transthoracic echocardiographic images with significant and limited overlap taken from apical window that is robust to the noise and intensity variation in ultrasound images. The algorithm estimates the translational and rotational components of the rigid transform through an iterative process and requires an initial approximation of the rotation and translation limits. We perform registration in two ways: the image-based registration computes the transform to align the end-diastolic frame of the apical nonstandard image to the apical standard image and applies the same transform to all frames of the cardiac cycle, whereas the mask-based registration approach uses the binary masks of the left ventricle in the same way. The SMC and exhaustive search (EX) algorithms were evaluated for 4D temporal sequences recorded from 7 volunteers who participated in a study conducted at the Mazankowski Alberta Heart Institute. The evaluations demonstrate that the mask-based approach of the accelerated SMC yielded a Dice score value of 0.819 +/- 0.045 for the left ventricle and gained 16.7x speedup compared to the CPU version of the SMC algorithm.
Neural Implicit Surface Reconstruction for Freehand 3D Ultrasound Volumetric Point Clouds with Geometric Constraints
Jan 13, 2024
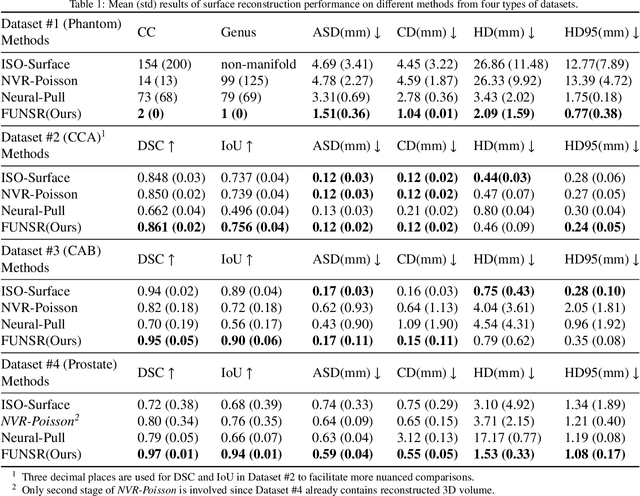
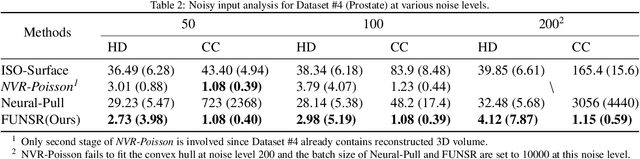
Abstract:Three-dimensional (3D) freehand ultrasound (US) is a widely used imaging modality that allows non-invasive imaging of medical anatomy without radiation exposure. The freehand 3D US surface reconstruction is vital to acquire the accurate anatomical structures needed for modeling, registration, and visualization. However, the currently used traditional methods cannot produce a high-quality surface due to imaging noise and connectivity issues in US. Although the deep learning-based approaches exhibiting the improvements in smoothness, continuity and resolution, the investigation into freehand 3D US remains limited. In this study, we introduce a self-supervised neural implicit surface reconstruction method to learn the signed distance functions (SDFs) from freehand 3D US volumetric point clouds. In particular, our method iteratively learns the SDFs by moving the 3D queries sampled around the point clouds to approximate the surface with the assistance of two novel geometric constraints. We assess our method on the three imaging systems, using twenty-three shapes that include six distinct anthropomorphic phantoms datasets and seventeen in vivo carotid artery datasets. Experimental results on phantoms outperform the existing approach, with a 67% reduction in Chamfer distance, 60% in Hausdorff distance, and 61% in Average absolute distance. Furthermore, our method achieves a 0.92 Dice score on the in vivo datasets and demonstrates great clinical potential.
Efficient automatic segmentation for multi-level pulmonary arteries: The PARSE challenge
Apr 07, 2023



Abstract:Efficient automatic segmentation of multi-level (i.e. main and branch) pulmonary arteries (PA) in CTPA images plays a significant role in clinical applications. However, most existing methods concentrate only on main PA or branch PA segmentation separately and ignore segmentation efficiency. Besides, there is no public large-scale dataset focused on PA segmentation, which makes it highly challenging to compare the different methods. To benchmark multi-level PA segmentation algorithms, we organized the first \textbf{P}ulmonary \textbf{AR}tery \textbf{SE}gmentation (PARSE) challenge. On the one hand, we focus on both the main PA and the branch PA segmentation. On the other hand, for better clinical application, we assign the same score weight to segmentation efficiency (mainly running time and GPU memory consumption during inference) while ensuring PA segmentation accuracy. We present a summary of the top algorithms and offer some suggestions for efficient and accurate multi-level PA automatic segmentation. We provide the PARSE challenge as open-access for the community to benchmark future algorithm developments at \url{https://parse2022.grand-challenge.org/Parse2022/}.
Biomedical image analysis competitions: The state of current participation practice
Dec 16, 2022Abstract:The number of international benchmarking competitions is steadily increasing in various fields of machine learning (ML) research and practice. So far, however, little is known about the common practice as well as bottlenecks faced by the community in tackling the research questions posed. To shed light on the status quo of algorithm development in the specific field of biomedical imaging analysis, we designed an international survey that was issued to all participants of challenges conducted in conjunction with the IEEE ISBI 2021 and MICCAI 2021 conferences (80 competitions in total). The survey covered participants' expertise and working environments, their chosen strategies, as well as algorithm characteristics. A median of 72% challenge participants took part in the survey. According to our results, knowledge exchange was the primary incentive (70%) for participation, while the reception of prize money played only a minor role (16%). While a median of 80 working hours was spent on method development, a large portion of participants stated that they did not have enough time for method development (32%). 25% perceived the infrastructure to be a bottleneck. Overall, 94% of all solutions were deep learning-based. Of these, 84% were based on standard architectures. 43% of the respondents reported that the data samples (e.g., images) were too large to be processed at once. This was most commonly addressed by patch-based training (69%), downsampling (37%), and solving 3D analysis tasks as a series of 2D tasks. K-fold cross-validation on the training set was performed by only 37% of the participants and only 50% of the participants performed ensembling based on multiple identical models (61%) or heterogeneous models (39%). 48% of the respondents applied postprocessing steps.
Unsupervised diffeomorphic cardiac image registration using parameterization of the deformation field
Aug 28, 2022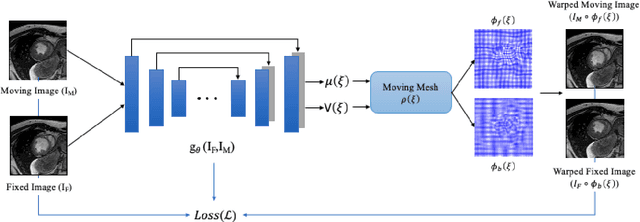


Abstract:This study proposes an end-to-end unsupervised diffeomorphic deformable registration framework based on moving mesh parameterization. Using this parameterization, a deformation field can be modeled with its transformation Jacobian determinant and curl of end velocity field. The new model of the deformation field has three important advantages; firstly, it relaxes the need for an explicit regularization term and the corresponding weight in the cost function. The smoothness is implicitly embedded in the solution which results in a physically plausible deformation field. Secondly, it guarantees diffeomorphism through explicit constraints applied to the transformation Jacobian determinant to keep it positive. Finally, it is suitable for cardiac data processing, since the nature of this parameterization is to define the deformation field in terms of the radial and rotational components. The effectiveness of the algorithm is investigated by evaluating the proposed method on three different data sets including 2D and 3D cardiac MRI scans. The results demonstrate that the proposed framework outperforms existing learning-based and non-learning-based methods while generating diffeomorphic transformations.
A training-free recursive multiresolution framework for diffeomorphic deformable image registration
Feb 01, 2022



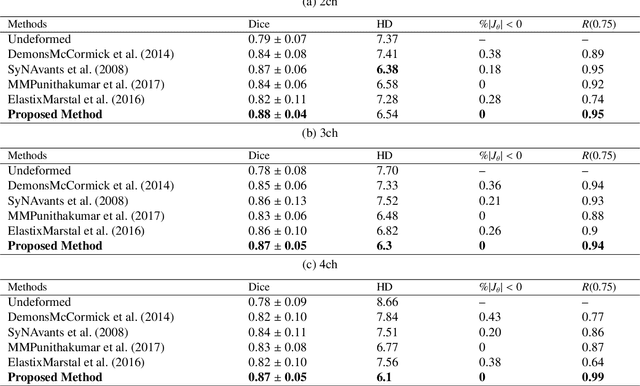
Abstract:Diffeomorphic deformable image registration is one of the crucial tasks in medical image analysis, which aims to find a unique transformation while preserving the topology and invertibility of the transformation. Deep convolutional neural networks (CNNs) have yielded well-suited approaches for image registration by learning the transformation priors from a large dataset. The improvement in the performance of these methods is related to their ability to learn information from several sample medical images that are difficult to obtain and bias the framework to the specific domain of data. In this paper, we propose a novel diffeomorphic training-free approach; this is built upon the principle of an ordinary differential equation. Our formulation yields an Euler integration type recursive scheme to estimate the changes of spatial transformations between the fixed and the moving image pyramids at different resolutions. The proposed architecture is simple in design. The moving image is warped successively at each resolution and finally aligned to the fixed image; this procedure is recursive in a way that at each resolution, a fully convolutional network (FCN) models a progressive change of deformation for the current warped image. The entire system is end-to-end and optimized for each pair of images from scratch. In comparison to learning-based methods, the proposed method neither requires a dedicated training set nor suffers from any training bias. We evaluate our method on three cardiac image datasets. The evaluation results demonstrate that the proposed method achieves state-of-the-art registration accuracy while maintaining desirable diffeomorphic properties.
MyoPS: A Benchmark of Myocardial Pathology Segmentation Combining Three-Sequence Cardiac Magnetic Resonance Images
Jan 10, 2022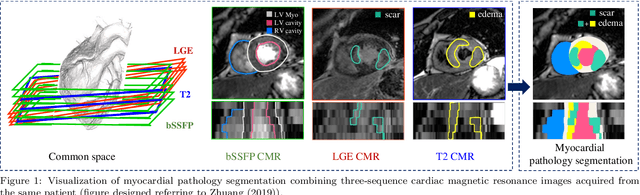
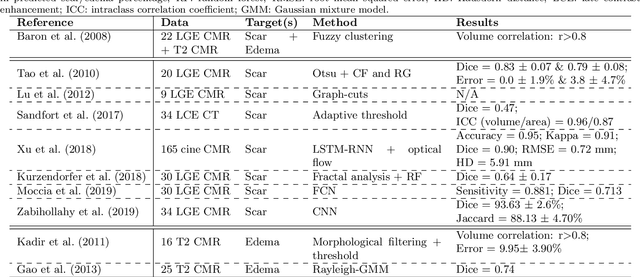
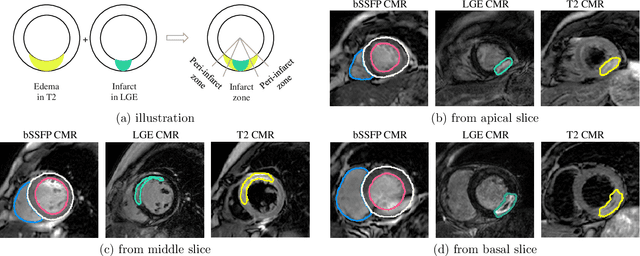
Abstract:Assessment of myocardial viability is essential in diagnosis and treatment management of patients suffering from myocardial infarction, and classification of pathology on myocardium is the key to this assessment. This work defines a new task of medical image analysis, i.e., to perform myocardial pathology segmentation (MyoPS) combining three-sequence cardiac magnetic resonance (CMR) images, which was first proposed in the MyoPS challenge, in conjunction with MICCAI 2020. The challenge provided 45 paired and pre-aligned CMR images, allowing algorithms to combine the complementary information from the three CMR sequences for pathology segmentation. In this article, we provide details of the challenge, survey the works from fifteen participants and interpret their methods according to five aspects, i.e., preprocessing, data augmentation, learning strategy, model architecture and post-processing. In addition, we analyze the results with respect to different factors, in order to examine the key obstacles and explore potential of solutions, as well as to provide a benchmark for future research. We conclude that while promising results have been reported, the research is still in the early stage, and more in-depth exploration is needed before a successful application to the clinics. Note that MyoPS data and evaluation tool continue to be publicly available upon registration via its homepage (www.sdspeople.fudan.edu.cn/zhuangxiahai/0/myops20/).
A New Semi-Automated Algorithm for Volumetric Segmentation of the Left Ventricle in Temporal 3D Echocardiography Sequences
Sep 03, 2021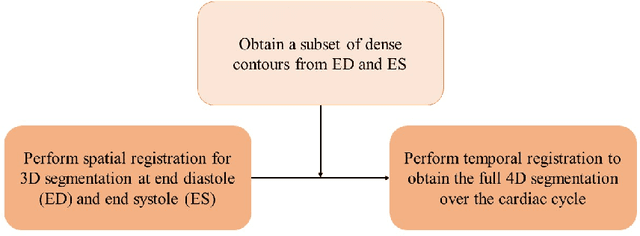
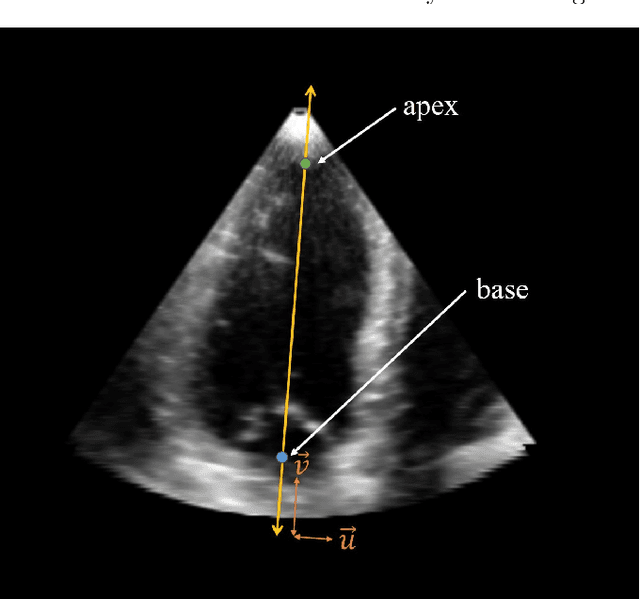
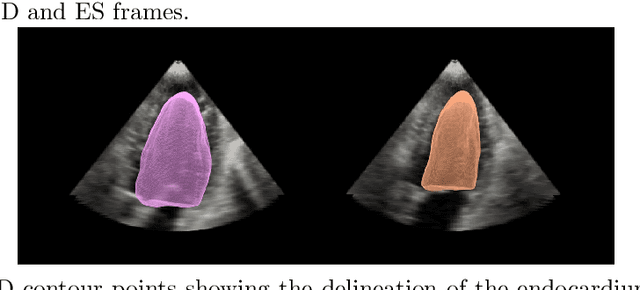
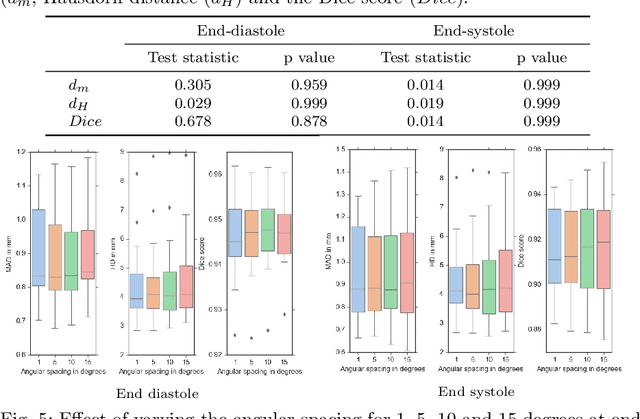
Abstract:Purpose: Echocardiography is commonly used as a non-invasive imaging tool in clinical practice for the assessment of cardiac function. However, delineation of the left ventricle is challenging due to the inherent properties of ultrasound imaging, such as the presence of speckle noise and the low signal-to-noise ratio. Methods: We propose a semi-automated segmentation algorithm for the delineation of the left ventricle in temporal 3D echocardiography sequences. The method requires minimal user interaction and relies on a diffeomorphic registration approach. Advantages of the method include no dependence on prior geometrical information, training data, or registration from an atlas. Results: The method was evaluated using three-dimensional ultrasound scan sequences from 18 patients from the Mazankowski Alberta Heart Institute, Edmonton, Canada, and compared to manual delineations provided by an expert cardiologist and four other registration algorithms. The segmentation approach yielded the following results over the cardiac cycle: a mean absolute difference of 1.01 (0.21) mm, a Hausdorff distance of 4.41 (1.43) mm, and a Dice overlap score of 0.93 (0.02). Conclusions: The method performed well compared to the four other registration algorithms.
* 22 pages, 8 figures
Fully Automated Left Atrium Segmentation from Anatomical Cine Long-axis MRI Sequences using Deep Convolutional Neural Network with Unscented Kalman Filter
Sep 28, 2020
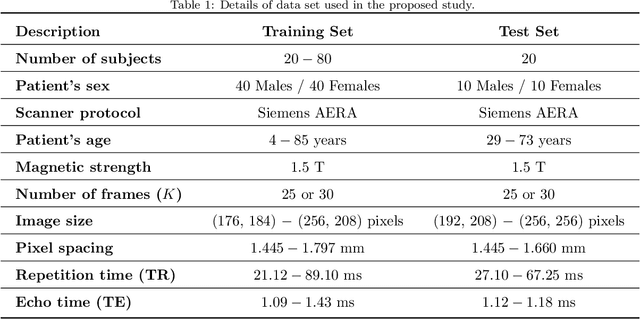

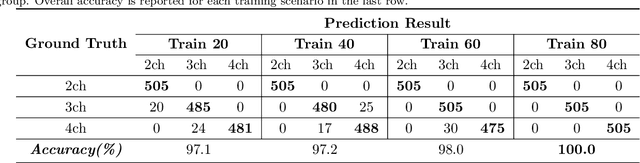
Abstract:This study proposes a fully automated approach for the left atrial segmentation from routine cine long-axis cardiac magnetic resonance image sequences using deep convolutional neural networks and Bayesian filtering. The proposed approach consists of a classification network that automatically detects the type of long-axis sequence and three different convolutional neural network models followed by unscented Kalman filtering (UKF) that delineates the left atrium. Instead of training and predicting all long-axis sequence types together, the proposed approach first identifies the image sequence type as to 2, 3 and 4 chamber views, and then performs prediction based on neural nets trained for that particular sequence type. The datasets were acquired retrospectively and ground truth manual segmentation was provided by an expert radiologist. In addition to neural net based classification and segmentation, another neural net is trained and utilized to select image sequences for further processing using UKF to impose temporal consistency over cardiac cycle. A cyclic dynamic model with time-varying angular frequency is introduced in UKF to characterize the variations in cardiac motion during image scanning. The proposed approach was trained and evaluated separately with varying amount of training data with images acquired from 20, 40, 60 and 80 patients. Evaluations over 1515 images with equal number of images from each chamber group acquired from an additional 20 patients demonstrated that the proposed model outperformed state-of-the-art and yielded a mean Dice coefficient value of 94.1%, 93.7% and 90.1% for 2, 3 and 4-chamber sequences, respectively, when trained with datasets from 80 patients.
Fully automated deep learning based segmentation of normal, infarcted and edema regions from multiple cardiac MRI sequences
Aug 18, 2020
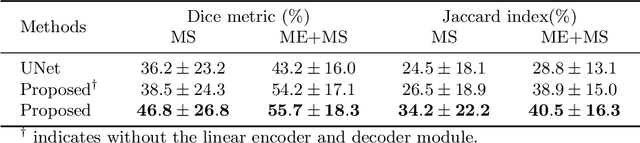


Abstract:Myocardial characterization is essential for patients with myocardial infarction and other myocardial diseases, and the assessment is often performed using cardiac magnetic resonance (CMR) sequences. In this study, we propose a fully automated approach using deep convolutional neural networks (CNN) for cardiac pathology segmentation, including left ventricular (LV) blood pool, right ventricular blood pool, LV normal myocardium, LV myocardial edema (ME) and LV myocardial scars (MS). The input to the network consists of three CMR sequences, namely, late gadolinium enhancement (LGE), T2 and balanced steady state free precession (bSSFP). The proposed approach utilized the data provided by the MyoPS challenge hosted by MICCAI 2020 in conjunction with STACOM. The training set for the CNN model consists of images acquired from 25 cases, and the gold standard labels are provided by trained raters and validated by radiologists. The proposed approach introduces a data augmentation module, linear encoder and decoder module and a network module to increase the number of training samples and improve the prediction accuracy for LV ME and MS. The proposed approach is evaluated by the challenge organizers with a test set including 20 cases and achieves a mean dice score of $46.8\%$ for LV MS and $55.7\%$ for LV ME+MS
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge