Ameneh Sheikhjafari
Unsupervised diffeomorphic cardiac image registration using parameterization of the deformation field
Aug 28, 2022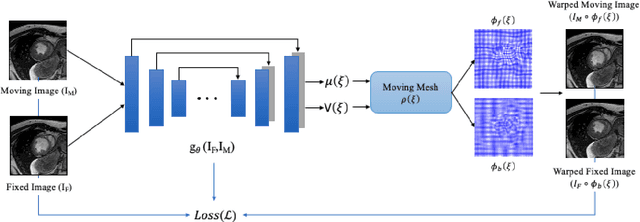

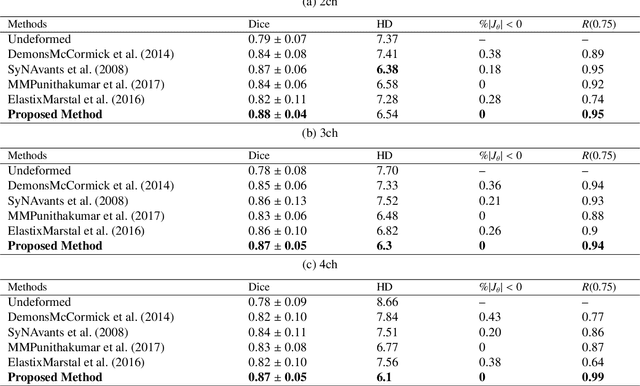

Abstract:This study proposes an end-to-end unsupervised diffeomorphic deformable registration framework based on moving mesh parameterization. Using this parameterization, a deformation field can be modeled with its transformation Jacobian determinant and curl of end velocity field. The new model of the deformation field has three important advantages; firstly, it relaxes the need for an explicit regularization term and the corresponding weight in the cost function. The smoothness is implicitly embedded in the solution which results in a physically plausible deformation field. Secondly, it guarantees diffeomorphism through explicit constraints applied to the transformation Jacobian determinant to keep it positive. Finally, it is suitable for cardiac data processing, since the nature of this parameterization is to define the deformation field in terms of the radial and rotational components. The effectiveness of the algorithm is investigated by evaluating the proposed method on three different data sets including 2D and 3D cardiac MRI scans. The results demonstrate that the proposed framework outperforms existing learning-based and non-learning-based methods while generating diffeomorphic transformations.
A training-free recursive multiresolution framework for diffeomorphic deformable image registration
Feb 01, 2022



Abstract:Diffeomorphic deformable image registration is one of the crucial tasks in medical image analysis, which aims to find a unique transformation while preserving the topology and invertibility of the transformation. Deep convolutional neural networks (CNNs) have yielded well-suited approaches for image registration by learning the transformation priors from a large dataset. The improvement in the performance of these methods is related to their ability to learn information from several sample medical images that are difficult to obtain and bias the framework to the specific domain of data. In this paper, we propose a novel diffeomorphic training-free approach; this is built upon the principle of an ordinary differential equation. Our formulation yields an Euler integration type recursive scheme to estimate the changes of spatial transformations between the fixed and the moving image pyramids at different resolutions. The proposed architecture is simple in design. The moving image is warped successively at each resolution and finally aligned to the fixed image; this procedure is recursive in a way that at each resolution, a fully convolutional network (FCN) models a progressive change of deformation for the current warped image. The entire system is end-to-end and optimized for each pair of images from scratch. In comparison to learning-based methods, the proposed method neither requires a dedicated training set nor suffers from any training bias. We evaluate our method on three cardiac image datasets. The evaluation results demonstrate that the proposed method achieves state-of-the-art registration accuracy while maintaining desirable diffeomorphic properties.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge