Lawrence H. Le
Accelerated 3D-3D rigid registration of echocardiographic images obtained from apical window using particle filter
Apr 28, 2025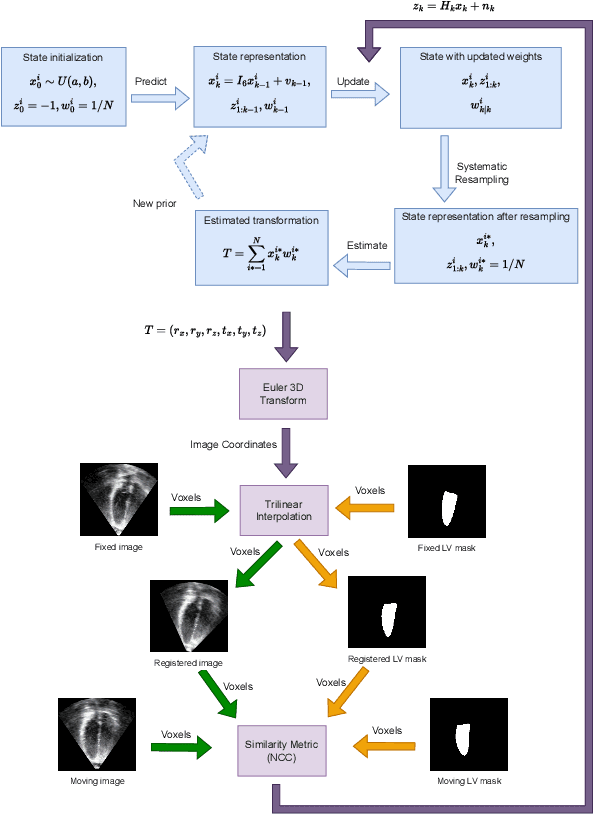



Abstract:The perfect alignment of 3D echocardiographic images captured from various angles has improved image quality and broadened the field of view. This study proposes an accelerated sequential Monte Carlo (SMC) algorithm for 3D-3D rigid registration of transthoracic echocardiographic images with significant and limited overlap taken from apical window that is robust to the noise and intensity variation in ultrasound images. The algorithm estimates the translational and rotational components of the rigid transform through an iterative process and requires an initial approximation of the rotation and translation limits. We perform registration in two ways: the image-based registration computes the transform to align the end-diastolic frame of the apical nonstandard image to the apical standard image and applies the same transform to all frames of the cardiac cycle, whereas the mask-based registration approach uses the binary masks of the left ventricle in the same way. The SMC and exhaustive search (EX) algorithms were evaluated for 4D temporal sequences recorded from 7 volunteers who participated in a study conducted at the Mazankowski Alberta Heart Institute. The evaluations demonstrate that the mask-based approach of the accelerated SMC yielded a Dice score value of 0.819 +/- 0.045 for the left ventricle and gained 16.7x speedup compared to the CPU version of the SMC algorithm.
Neural Implicit Surface Reconstruction for Freehand 3D Ultrasound Volumetric Point Clouds with Geometric Constraints
Jan 13, 2024
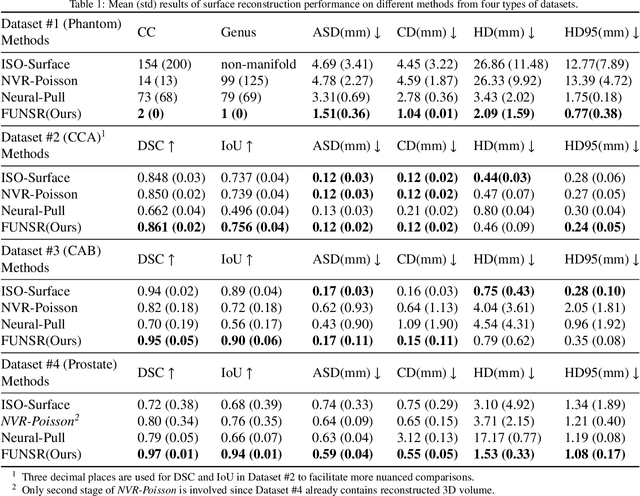
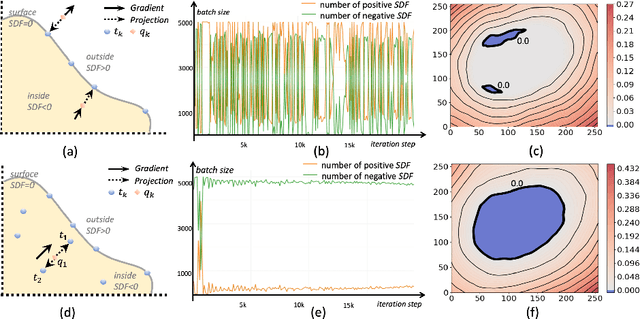
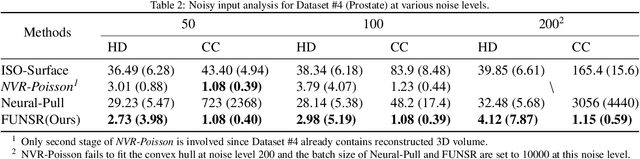
Abstract:Three-dimensional (3D) freehand ultrasound (US) is a widely used imaging modality that allows non-invasive imaging of medical anatomy without radiation exposure. The freehand 3D US surface reconstruction is vital to acquire the accurate anatomical structures needed for modeling, registration, and visualization. However, the currently used traditional methods cannot produce a high-quality surface due to imaging noise and connectivity issues in US. Although the deep learning-based approaches exhibiting the improvements in smoothness, continuity and resolution, the investigation into freehand 3D US remains limited. In this study, we introduce a self-supervised neural implicit surface reconstruction method to learn the signed distance functions (SDFs) from freehand 3D US volumetric point clouds. In particular, our method iteratively learns the SDFs by moving the 3D queries sampled around the point clouds to approximate the surface with the assistance of two novel geometric constraints. We assess our method on the three imaging systems, using twenty-three shapes that include six distinct anthropomorphic phantoms datasets and seventeen in vivo carotid artery datasets. Experimental results on phantoms outperform the existing approach, with a 67% reduction in Chamfer distance, 60% in Hausdorff distance, and 61% in Average absolute distance. Furthermore, our method achieves a 0.92 Dice score on the in vivo datasets and demonstrates great clinical potential.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge