Yuqian Chen
Multi-dimensional Assessment and Explainable Feedback for Counselor Responses to Client Resistance in Text-based Counseling with LLMs
Feb 25, 2026Abstract:Effectively addressing client resistance is a sophisticated clinical skill in psychological counseling, yet practitioners often lack timely and scalable supervisory feedback to refine their approaches. Although current NLP research has examined overall counseling quality and general therapeutic skills, it fails to provide granular evaluations of high-stakes moments where clients exhibit resistance. In this work, we present a comprehensive pipeline for the multi-dimensional evaluation of human counselors' interventions specifically targeting client resistance in text-based therapy. We introduce a theory-driven framework that decomposes counselor responses into four distinct communication mechanisms. Leveraging this framework, we curate and share an expert-annotated dataset of real-world counseling excerpts, pairing counselor-client interactions with professional ratings and explanatory rationales. Using this data, we perform full-parameter instruction tuning on a Llama-3.1-8B-Instruct backbone to model fine-grained evaluative judgments of response quality and generate explanations underlying. Experimental results show that our approach can effectively distinguish the quality of different communication mechanisms (77-81% F1), substantially outperforming GPT-4o and Claude-3.5-Sonnet (45-59% F1). Moreover, the model produces high-quality explanations that closely align with expert references and receive near-ceiling ratings from human experts (2.8-2.9/3.0). A controlled experiment with 43 counselors further confirms that receiving these AI-generated feedback significantly improves counselors' ability to respond effectively to client resistance.
RECAP: Resistance Capture in Text-based Mental Health Counseling with Large Language Models
Jan 21, 2026Abstract:Recognizing and navigating client resistance is critical for effective mental health counseling, yet detecting such behaviors is particularly challenging in text-based interactions. Existing NLP approaches oversimplify resistance categories, ignore the sequential dynamics of therapeutic interventions, and offer limited interpretability. To address these limitations, we propose PsyFIRE, a theoretically grounded framework capturing 13 fine-grained resistance behaviors alongside collaborative interactions. Based on PsyFIRE, we construct the ClientResistance corpus with 23,930 annotated utterances from real-world Chinese text-based counseling, each supported by context-specific rationales. Leveraging this dataset, we develop RECAP, a two-stage framework that detects resistance and fine-grained resistance types with explanations. RECAP achieves 91.25% F1 for distinguishing collaboration and resistance and 66.58% macro-F1 for fine-grained resistance categories classification, outperforming leading prompt-based LLM baselines by over 20 points. Applied to a separate counseling dataset and a pilot study with 62 counselors, RECAP reveals the prevalence of resistance, its negative impact on therapeutic relationships and demonstrates its potential to improve counselors' understanding and intervention strategies.
Bridging Modalities: Joint Synthesis and Registration Framework for Aligning Diffusion MRI with T1-Weighted Images
Jan 16, 2026Abstract:Multimodal image registration between diffusion MRI (dMRI) and T1-weighted (T1w) MRI images is a critical step for aligning diffusion-weighted imaging (DWI) data with structural anatomical space. Traditional registration methods often struggle to ensure accuracy due to the large intensity differences between diffusion data and high-resolution anatomical structures. This paper proposes an unsupervised registration framework based on a generative registration network, which transforms the original multimodal registration problem between b0 and T1w images into a unimodal registration task between a generated image and the real T1w image. This effectively reduces the complexity of cross-modal registration. The framework first employs an image synthesis model to generate images with T1w-like contrast, and then learns a deformation field from the generated image to the fixed T1w image. The registration network jointly optimizes local structural similarity and cross-modal statistical dependency to improve deformation estimation accuracy. Experiments conducted on two independent datasets demonstrate that the proposed method outperforms several state-of-the-art approaches in multimodal registration tasks.
PsyCLIENT: Client Simulation via Conversational Trajectory Modeling for Trainee Practice and Model Evaluation in Mental Health Counseling
Jan 12, 2026Abstract:LLM-based client simulation has emerged as a promising tool for training novice counselors and evaluating automated counseling systems. However, existing client simulation approaches face three key challenges: (1) limited diversity and realism in client profiles, (2) the lack of a principled framework for modeling realistic client behaviors, and (3) a scarcity in Chinese-language settings. To address these limitations, we propose PsyCLIENT, a novel simulation framework grounded in conversational trajectory modeling. By conditioning LLM generation on predefined real-world trajectories that incorporate explicit behavior labels and content constraints, our approach ensures diverse and realistic interactions. We further introduce PsyCLIENT-CP, the first open-source Chinese client profile dataset, covering 60 distinct counseling topics. Comprehensive evaluations involving licensed professional counselors demonstrate that PsyCLIENT significantly outperforms baselines in terms of authenticity and training effectiveness. Notably, the simulated clients are nearly indistinguishable from human clients, achieving an about 95\% expert confusion rate in discrimination tasks. These findings indicate that conversational trajectory modeling effectively bridges the gap between theoretical client profiles and dynamic, realistic simulations, offering a robust solution for mental health education and research. Code and data will be released to facilitate future research in mental health counseling.
Cross-Population White Matter Atlas Creation for Concurrent Mapping of Brain Connections in Neonates and Adults with Diffusion MRI Tractography
Dec 23, 2025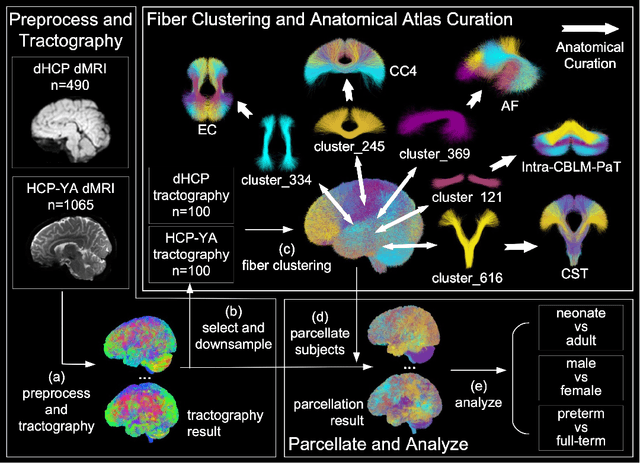
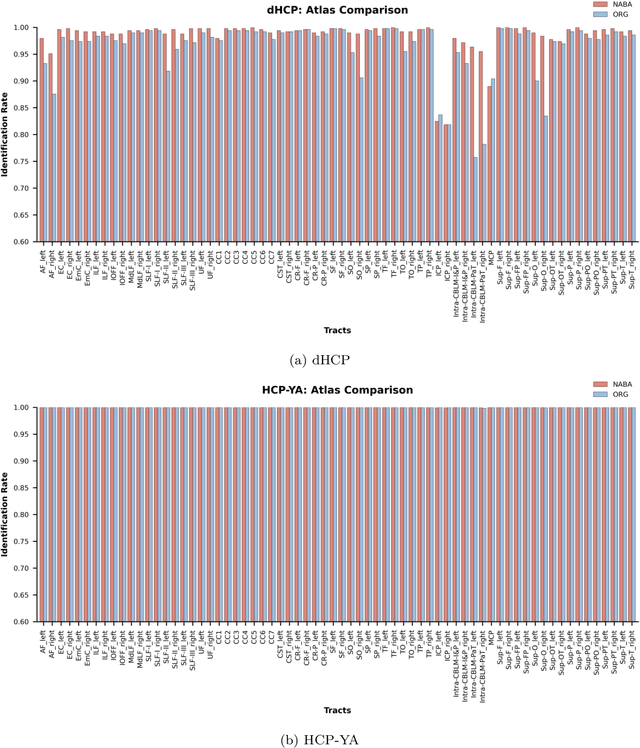
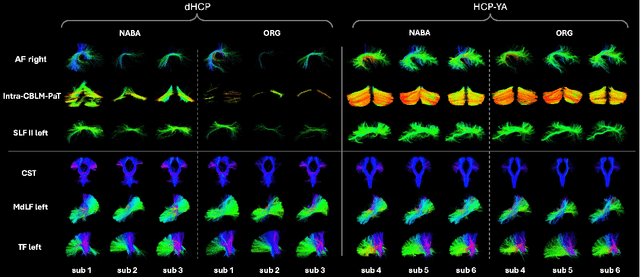
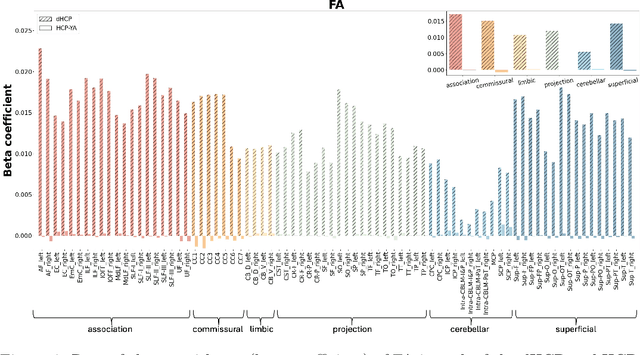
Abstract:Comparing white matter (WM) connections between adults and neonates using diffusion MRI (dMRI) can advance our understanding of typical brain development and potential biomarkers for neurological disorders. However, existing WM atlases are population-specific (adult or neonatal) and reside in separate spaces, preventing direct cross-population comparisons. A unified WM atlas spanning both neonates and adults is still lacking. In this study, we propose a neonatal/adult brain atlas (NABA), a WM tractography atlas built from dMRI data of both neonates and adults. NABA is constructed using a robust, data-driven fiber clustering pipeline, enabling group-wise WM atlasing across populations despite substantial anatomical variability. The atlas provides a standardized template for WM parcellation, allowing direct comparison of WM tracts between neonates and adults. Using NABA, we conduct four analyses: (1) evaluating the feasibility of joint WM mapping across populations, (2) characterizing WM development across neonatal ages relative to adults, (3) assessing sex-related differences in neonatal WM development, and (4) examining the effects of preterm birth. Our results show that NABA robustly identifies WM tracts in both populations. We observe rapid fractional anisotropy (FA) development in long-range association tracts, including the arcuate fasciculus and superior longitudinal fasciculus II, whereas intra-cerebellar tracts develop more slowly. Neonatal females exhibit faster overall FA development than males. Although preterm neonates show lower overall FA development rates, they demonstrate relatively higher FA growth in specific tracts, including the corticospinal tract, corona radiata-pontine pathway, and intracerebellar tracts. These findings demonstrate that NABA is a useful tool for investigating WM development across neonates and adults.
DeepMultiConnectome: Deep Multi-Task Prediction of Structural Connectomes Directly from Diffusion MRI Tractography
May 27, 2025



Abstract:Diffusion MRI (dMRI) tractography enables in vivo mapping of brain structural connections, but traditional connectome generation is time-consuming and requires gray matter parcellation, posing challenges for large-scale studies. We introduce DeepMultiConnectome, a deep-learning model that predicts structural connectomes directly from tractography, bypassing the need for gray matter parcellation while supporting multiple parcellation schemes. Using a point-cloud-based neural network with multi-task learning, the model classifies streamlines according to their connected regions across two parcellation schemes, sharing a learned representation. We train and validate DeepMultiConnectome on tractography from the Human Connectome Project Young Adult dataset ($n = 1000$), labeled with an 84 and 164 region gray matter parcellation scheme. DeepMultiConnectome predicts multiple structural connectomes from a whole-brain tractogram containing 3 million streamlines in approximately 40 seconds. DeepMultiConnectome is evaluated by comparing predicted connectomes with traditional connectomes generated using the conventional method of labeling streamlines using a gray matter parcellation. The predicted connectomes are highly correlated with traditionally generated connectomes ($r = 0.992$ for an 84-region scheme; $r = 0.986$ for a 164-region scheme) and largely preserve network properties. A test-retest analysis of DeepMultiConnectome demonstrates reproducibility comparable to traditionally generated connectomes. The predicted connectomes perform similarly to traditionally generated connectomes in predicting age and cognitive function. Overall, DeepMultiConnectome provides a scalable, fast model for generating subject-specific connectomes across multiple parcellation schemes.
A Multimodal Deep Learning Approach for White Matter Shape Prediction in Diffusion MRI Tractography
Apr 25, 2025Abstract:Shape measures have emerged as promising descriptors of white matter tractography, offering complementary insights into anatomical variability and associations with cognitive and clinical phenotypes. However, conventional methods for computing shape measures are computationally expensive and time-consuming for large-scale datasets due to reliance on voxel-based representations. We propose Tract2Shape, a novel multimodal deep learning framework that leverages geometric (point cloud) and scalar (tabular) features to predict ten white matter tractography shape measures. To enhance model efficiency, we utilize a dimensionality reduction algorithm for the model to predict five primary shape components. The model is trained and evaluated on two independently acquired datasets, the HCP-YA dataset, and the PPMI dataset. We evaluate the performance of Tract2Shape by training and testing it on the HCP-YA dataset and comparing the results with state-of-the-art models. To further assess its robustness and generalization ability, we also test Tract2Shape on the unseen PPMI dataset. Tract2Shape outperforms SOTA deep learning models across all ten shape measures, achieving the highest average Pearson's r and the lowest nMSE on the HCP-YA dataset. The ablation study shows that both multimodal input and PCA contribute to performance gains. On the unseen testing PPMI dataset, Tract2Shape maintains a high Pearson's r and low nMSE, demonstrating strong generalizability in cross-dataset evaluation. Tract2Shape enables fast, accurate, and generalizable prediction of white matter shape measures from tractography data, supporting scalable analysis across datasets. This framework lays a promising foundation for future large-scale white matter shape analysis.
NTIRE 2025 Challenge on Day and Night Raindrop Removal for Dual-Focused Images: Methods and Results
Apr 19, 2025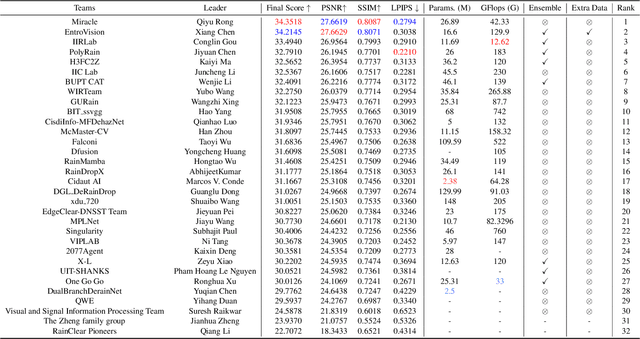
Abstract:This paper reviews the NTIRE 2025 Challenge on Day and Night Raindrop Removal for Dual-Focused Images. This challenge received a wide range of impressive solutions, which are developed and evaluated using our collected real-world Raindrop Clarity dataset. Unlike existing deraining datasets, our Raindrop Clarity dataset is more diverse and challenging in degradation types and contents, which includes day raindrop-focused, day background-focused, night raindrop-focused, and night background-focused degradations. This dataset is divided into three subsets for competition: 14,139 images for training, 240 images for validation, and 731 images for testing. The primary objective of this challenge is to establish a new and powerful benchmark for the task of removing raindrops under varying lighting and focus conditions. There are a total of 361 participants in the competition, and 32 teams submitting valid solutions and fact sheets for the final testing phase. These submissions achieved state-of-the-art (SOTA) performance on the Raindrop Clarity dataset. The project can be found at https://lixinustc.github.io/CVPR-NTIRE2025-RainDrop-Competition.github.io/.
DeepNuParc: A Novel Deep Clustering Framework for Fine-scale Parcellation of Brain Nuclei Using Diffusion MRI Tractography
Mar 10, 2025



Abstract:Brain nuclei are clusters of anatomically distinct neurons that serve as important hubs for processing and relaying information in various neural circuits. Fine-scale parcellation of the brain nuclei is vital for a comprehensive understanding of its anatomico-functional correlations. Diffusion MRI tractography is an advanced imaging technique that can estimate the brain's white matter structural connectivity to potentially reveal the topography of the nuclei of interest for studying its subdivisions. In this work, we present a deep clustering pipeline, namely DeepNuParc, to perform automated, fine-scale parcellation of brain nuclei using diffusion MRI tractography. First, we incorporate a newly proposed deep learning approach to enable accurate segmentation of the nuclei of interest directly on the dMRI data. Next, we design a novel streamline clustering-based structural connectivity feature for a robust representation of voxels within the nuclei. Finally, we improve the popular joint dimensionality reduction and k-means clustering approach to enable nuclei parcellation at a finer scale. We demonstrate DeepNuParc on two important brain structures, i.e. the amygdala and the thalamus, that are known to have multiple anatomically and functionally distinct nuclei subdivisions. Experimental results show that DeepNuParc enables consistent parcellation of the nuclei into multiple parcels across multiple subjects and achieves good correspondence with the widely used coarse-scale atlases. Our codes are available at https://github.com/HarlandZZC/deep_nuclei_parcellation.
MultiCo3D: Multi-Label Voxel Contrast for One-Shot Incremental Segmentation of 3D Neuroimages
Mar 09, 2025Abstract:3D neuroimages provide a comprehensive view of brain structure and function, aiding in precise localization and functional connectivity analysis. Segmentation of white matter (WM) tracts using 3D neuroimages is vital for understanding the brain's structural connectivity in both healthy and diseased states. One-shot Class Incremental Semantic Segmentation (OCIS) refers to effectively segmenting new (novel) classes using only a single sample while retaining knowledge of old (base) classes without forgetting. Voxel-contrastive OCIS methods adjust the feature space to alleviate the feature overlap problem between the base and novel classes. However, since WM tract segmentation is a multi-label segmentation task, existing single-label voxel contrastive-based methods may cause inherent contradictions. To address this, we propose a new multi-label voxel contrast framework called MultiCo3D for one-shot class incremental tract segmentation. Our method utilizes uncertainty distillation to preserve base tract segmentation knowledge while adjusting the feature space with multi-label voxel contrast to alleviate feature overlap when learning novel tracts and dynamically weighting multi losses to balance overall loss. We compare our method against several state-of-the-art (SOTA) approaches. The experimental results show that our method significantly enhances one-shot class incremental tract segmentation accuracy across five different experimental setups on HCP and Preto datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge