Hanchuan Peng
DINeuro: Distilling Knowledge from 2D Natural Images via Deformable Tubular Transferring Strategy for 3D Neuron Reconstruction
Oct 29, 2024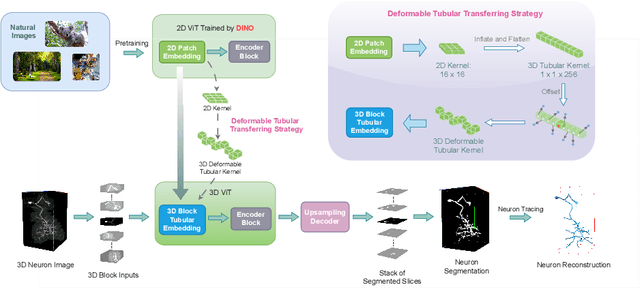
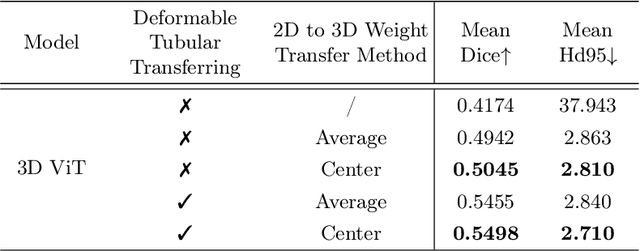
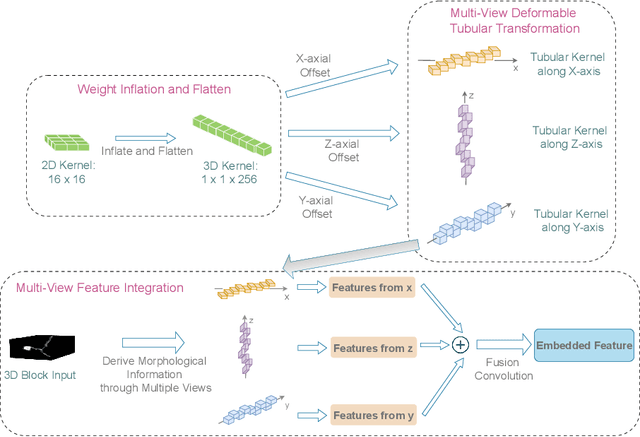
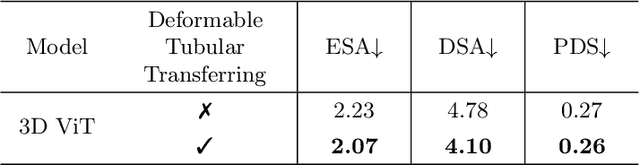
Abstract:Reconstructing neuron morphology from 3D light microscope imaging data is critical to aid neuroscientists in analyzing brain networks and neuroanatomy. With the boost from deep learning techniques, a variety of learning-based segmentation models have been developed to enhance the signal-to-noise ratio of raw neuron images as a pre-processing step in the reconstruction workflow. However, most existing models directly encode the latent representative features of volumetric neuron data but neglect their intrinsic morphological knowledge. To address this limitation, we design a novel framework that distills the prior knowledge from a 2D Vision Transformer pre-trained on extensive 2D natural images to facilitate neuronal morphological learning of our 3D Vision Transformer. To bridge the knowledge gap between the 2D natural image and 3D microscopic morphologic domains, we propose a deformable tubular transferring strategy that adapts the pre-trained 2D natural knowledge to the inherent tubular characteristics of neuronal structure in the latent embedding space. The experimental results on the Janelia dataset of the BigNeuron project demonstrate that our method achieves a segmentation performance improvement of 4.53% in mean Dice and 3.56% in mean 95% Hausdorff distance.
Boosting 3D Neuron Segmentation with 2D Vision Transformer Pre-trained on Natural Images
May 04, 2024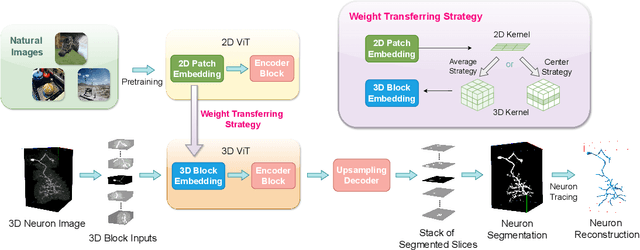
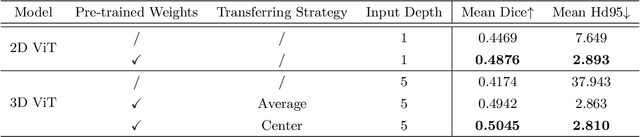
Abstract:Neuron reconstruction, one of the fundamental tasks in neuroscience, rebuilds neuronal morphology from 3D light microscope imaging data. It plays a critical role in analyzing the structure-function relationship of neurons in the nervous system. However, due to the scarcity of neuron datasets and high-quality SWC annotations, it is still challenging to develop robust segmentation methods for single neuron reconstruction. To address this limitation, we aim to distill the consensus knowledge from massive natural image data to aid the segmentation model in learning the complex neuron structures. Specifically, in this work, we propose a novel training paradigm that leverages a 2D Vision Transformer model pre-trained on large-scale natural images to initialize our Transformer-based 3D neuron segmentation model with a tailored 2D-to-3D weight transferring strategy. Our method builds a knowledge sharing connection between the abundant natural and the scarce neuron image domains to improve the 3D neuron segmentation ability in a data-efficiency manner. Evaluated on a popular benchmark, BigNeuron, our method enhances neuron segmentation performance by 8.71% over the model trained from scratch with the same amount of training samples.
Towards NeuroAI: Introducing Neuronal Diversity into Artificial Neural Networks
Jan 23, 2023
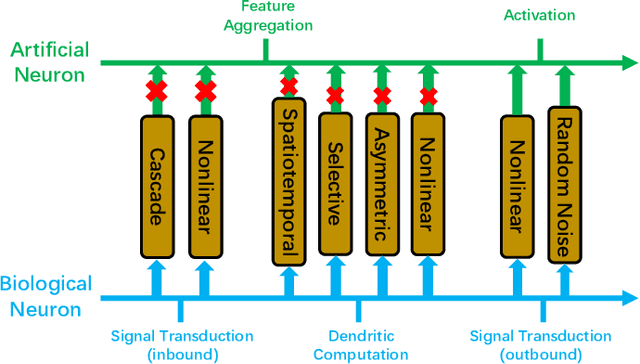
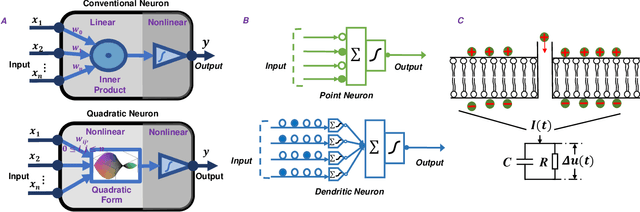
Abstract:Throughout history, the development of artificial intelligence, particularly artificial neural networks, has been open to and constantly inspired by the increasingly deepened understanding of the brain, such as the inspiration of neocognitron, which is the pioneering work of convolutional neural networks. Per the motives of the emerging field: NeuroAI, a great amount of neuroscience knowledge can help catalyze the next generation of AI by endowing a network with more powerful capabilities. As we know, the human brain has numerous morphologically and functionally different neurons, while artificial neural networks are almost exclusively built on a single neuron type. In the human brain, neuronal diversity is an enabling factor for all kinds of biological intelligent behaviors. Since an artificial network is a miniature of the human brain, introducing neuronal diversity should be valuable in terms of addressing those essential problems of artificial networks such as efficiency, interpretability, and memory. In this Primer, we first discuss the preliminaries of biological neuronal diversity and the characteristics of information transmission and processing in a biological neuron. Then, we review studies of designing new neurons for artificial networks. Next, we discuss what gains can neuronal diversity bring into artificial networks and exemplary applications in several important fields. Lastly, we discuss the challenges and future directions of neuronal diversity to explore the potential of NeuroAI.
Single Neuron Segmentation using Graph-based Global Reasoning with Auxiliary Skeleton Loss from 3D Optical Microscope Images
Jan 22, 2021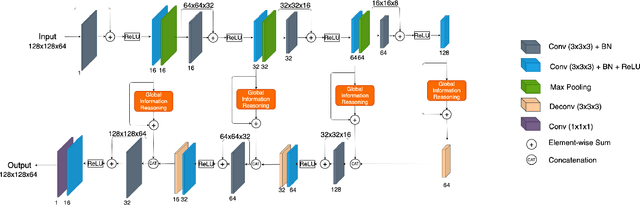

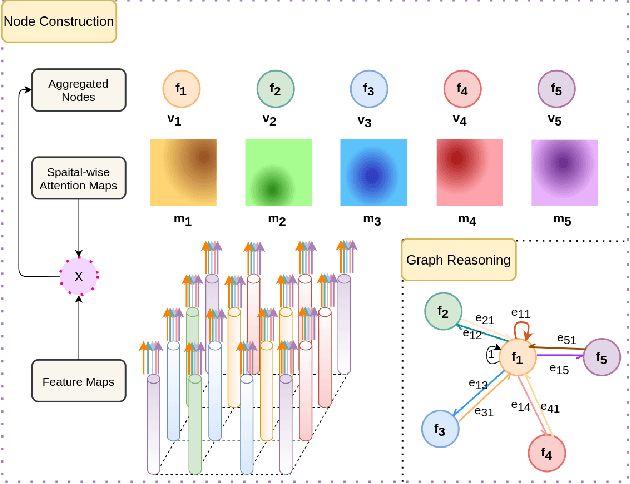

Abstract:One of the critical steps in improving accurate single neuron reconstruction from three-dimensional (3D) optical microscope images is the neuronal structure segmentation. However, they are always hard to segment due to the lack in quality. Despite a series of attempts to apply convolutional neural networks (CNNs) on this task, noise and disconnected gaps are still challenging to alleviate with the neglect of the non-local features of graph-like tubular neural structures. Hence, we present an end-to-end segmentation network by jointly considering the local appearance and the global geometry traits through graph reasoning and a skeleton-based auxiliary loss. The evaluation results on the Janelia dataset from the BigNeuron project demonstrate that our proposed method exceeds the counterpart algorithms in performance.
AIDE: Annotation-efficient deep learning for automatic medical image segmentation
Dec 14, 2020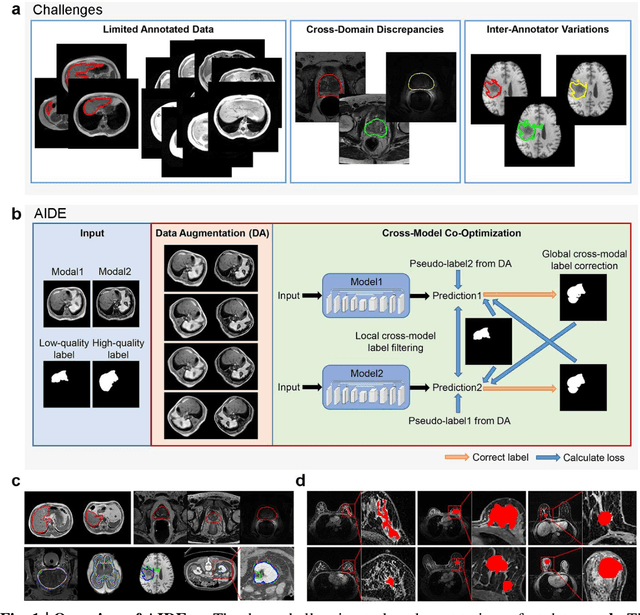
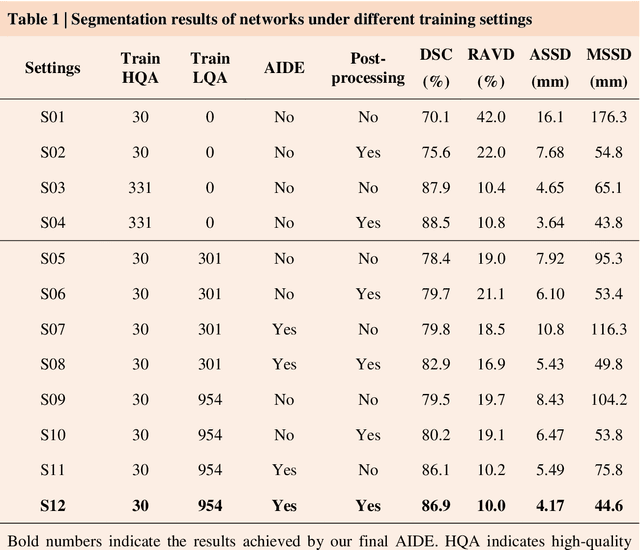
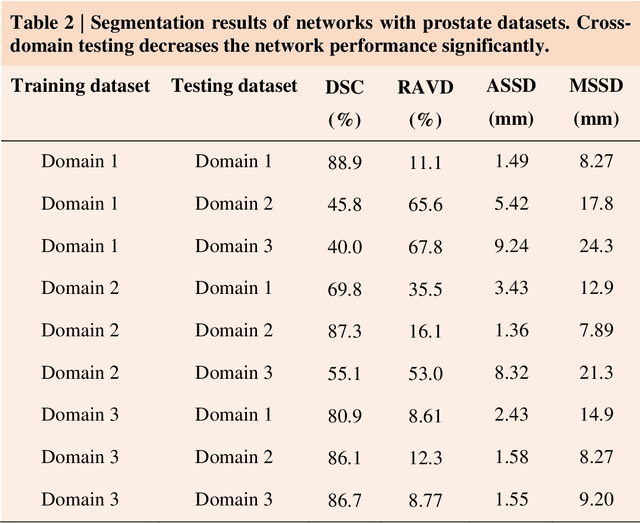
Abstract:Accurate image segmentation is crucial for medical imaging applications. The prevailing deep learning approaches typically rely on very large training datasets with high-quality manual annotations, which are often not available in medical imaging. We introduce Annotation-effIcient Deep lEarning (AIDE) to handle imperfect datasets with an elaborately designed cross-model self-correcting mechanism. AIDE improves the segmentation Dice scores of conventional deep learning models on open datasets possessing scarce or noisy annotations by up to 30%. For three clinical datasets containing 11,852 breast images of 872 patients from three medical centers, AIDE consistently produces segmentation maps comparable to those generated by the fully supervised counterparts as well as the manual annotations of independent radiologists by utilizing only 10% training annotations. Such a 10-fold improvement of efficiency in utilizing experts' labels has the potential to promote a wide range of biomedical applications.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge