Rongpin Wang
SUFFICIENT: A scan-specific unsupervised deep learning framework for high-resolution 3D isotropic fetal brain MRI reconstruction
May 26, 2025Abstract:High-quality 3D fetal brain MRI reconstruction from motion-corrupted 2D slices is crucial for clinical diagnosis. Reliable slice-to-volume registration (SVR)-based motion correction and super-resolution reconstruction (SRR) methods are essential. Deep learning (DL) has demonstrated potential in enhancing SVR and SRR when compared to conventional methods. However, it requires large-scale external training datasets, which are difficult to obtain for clinical fetal MRI. To address this issue, we propose an unsupervised iterative SVR-SRR framework for isotropic HR volume reconstruction. Specifically, SVR is formulated as a function mapping a 2D slice and a 3D target volume to a rigid transformation matrix, which aligns the slice to the underlying location in the target volume. The function is parameterized by a convolutional neural network, which is trained by minimizing the difference between the volume slicing at the predicted position and the input slice. In SRR, a decoding network embedded within a deep image prior framework is incorporated with a comprehensive image degradation model to produce the high-resolution (HR) volume. The deep image prior framework offers a local consistency prior to guide the reconstruction of HR volumes. By performing a forward degradation model, the HR volume is optimized by minimizing loss between predicted slices and the observed slices. Comprehensive experiments conducted on large-magnitude motion-corrupted simulation data and clinical data demonstrate the superior performance of the proposed framework over state-of-the-art fetal brain reconstruction frameworks.
2D and 3D CT Radiomic Features Performance Comparison in Characterization of Gastric Cancer: A Multi-center Study
Oct 29, 2022Abstract:Objective: Radiomics, an emerging tool for medical image analysis, is potential towards precisely characterizing gastric cancer (GC). Whether using one-slice 2D annotation or whole-volume 3D annotation remains a long-time debate, especially for heterogeneous GC. We comprehensively compared 2D and 3D radiomic features' representation and discrimination capacity regarding GC, via three tasks. Methods: Four-center 539 GC patients were retrospectively enrolled and divided into the training and validation cohorts. From 2D or 3D regions of interest (ROIs) annotated by radiologists, radiomic features were extracted respectively. Feature selection and model construction procedures were customed for each combination of two modalities (2D or 3D) and three tasks. Subsequently, six machine learning models (Model_2D^LNM, Model_3D^LNM; Model_2D^LVI, Model_3D^LVI; Model_2D^pT, Model_3D^pT) were derived and evaluated to reflect modalities' performances in characterizing GC. Furthermore, we performed an auxiliary experiment to assess modalities' performances when resampling spacing is different. Results: Regarding three tasks, the yielded areas under the curve (AUCs) were: Model_2D^LNM's 0.712 (95% confidence interval, 0.613-0.811), Model_3D^LNM's 0.680 (0.584-0.775); Model_2D^LVI's 0.677 (0.595-0.761), Model_3D^LVI's 0.615 (0.528-0.703); Model_2D^pT's 0.840 (0.779-0.901), Model_3D^pT's 0.813 (0.747-0.879). Moreover, the auxiliary experiment indicated that Models_2D are statistically more advantageous than Models3D with different resampling spacings. Conclusion: Models constructed with 2D radiomic features revealed comparable performances with those constructed with 3D features in characterizing GC. Significance: Our work indicated that time-saving 2D annotation would be the better choice in GC, and provided a related reference to further radiomics-based researches.
* Published in IEEE Journal of Biomedical and Health Informatics
AIDE: Annotation-efficient deep learning for automatic medical image segmentation
Dec 14, 2020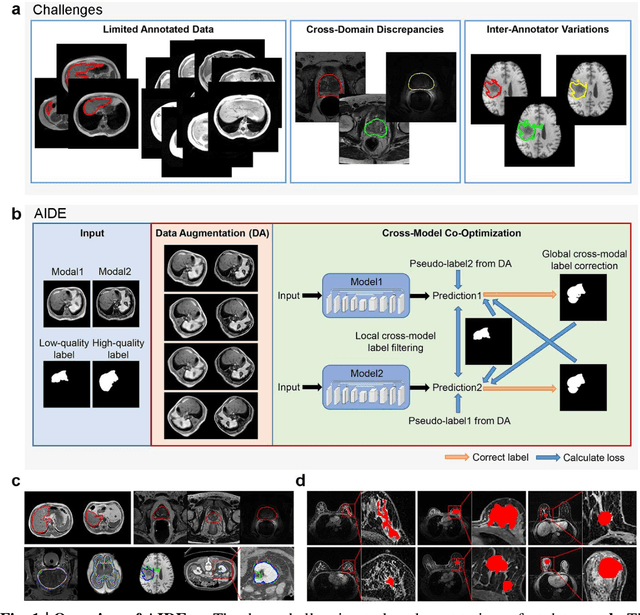
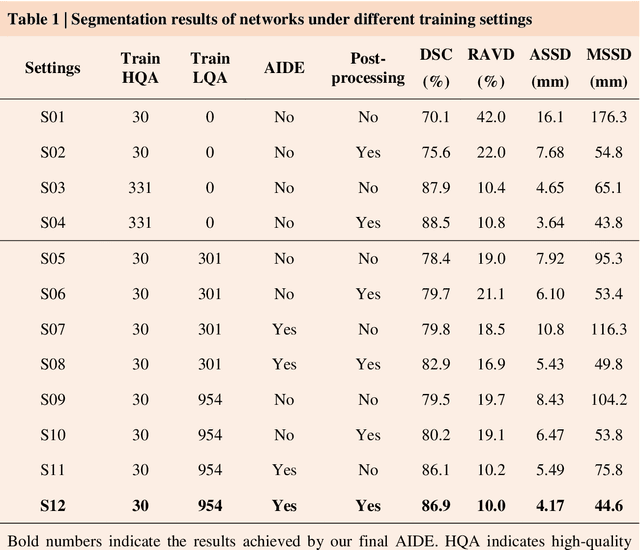
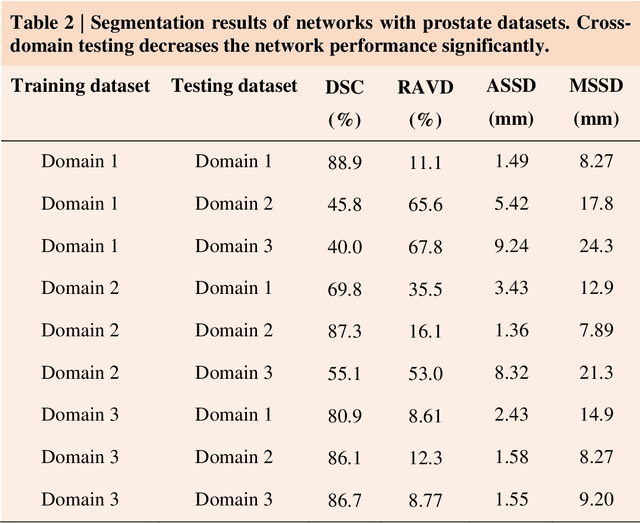
Abstract:Accurate image segmentation is crucial for medical imaging applications. The prevailing deep learning approaches typically rely on very large training datasets with high-quality manual annotations, which are often not available in medical imaging. We introduce Annotation-effIcient Deep lEarning (AIDE) to handle imperfect datasets with an elaborately designed cross-model self-correcting mechanism. AIDE improves the segmentation Dice scores of conventional deep learning models on open datasets possessing scarce or noisy annotations by up to 30%. For three clinical datasets containing 11,852 breast images of 872 patients from three medical centers, AIDE consistently produces segmentation maps comparable to those generated by the fully supervised counterparts as well as the manual annotations of independent radiologists by utilizing only 10% training annotations. Such a 10-fold improvement of efficiency in utilizing experts' labels has the potential to promote a wide range of biomedical applications.
A coarse-to-fine framework for unsupervised multi-contrast MR image deformable registration with dual consistency constraint
Aug 06, 2020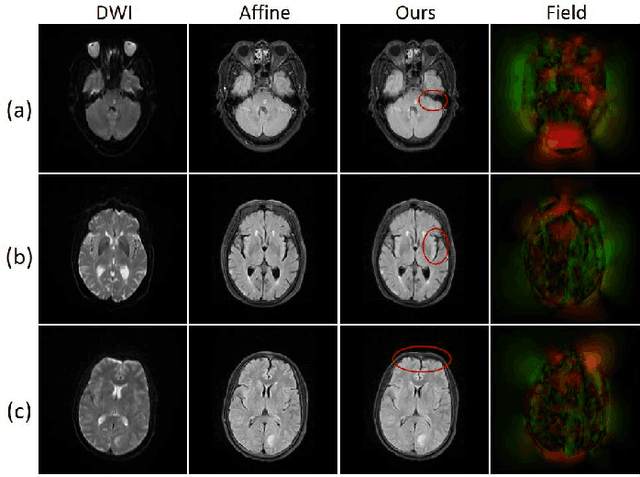
Abstract:Multi-contrast magnetic resonance (MR) image registration is essential in the clinic to achieve fast and accurate imaging-based disease diagnosis and treatment planning. Nevertheless, the efficiency and performance of the existing registration algorithms can still be improved. In this paper, we propose a novel unsupervised learning-based framework to achieve accurate and efficient multi-contrast MR image registrations. Specifically, an end-to-end coarse-to-fine network architecture consisting of affine and deformable transformations is designed to get rid of both the multi-step iteration process and the complex image preprocessing operations. Furthermore, a dual consistency constraint and a new prior knowledge-based loss function are developed to enhance the registration performances. The proposed method has been evaluated on a clinical dataset that consists of 555 cases, with encouraging performances achieved. Compared to the commonly utilized registration methods, including Voxelmorph, SyN, and LDDMM, the proposed method achieves the best registration performance with a Dice score of 0.826 in identifying stroke lesions. More robust performance in low-signal areas is also observed. With regards to the registration speed, our method is about 17 times faster than the most competitive method of SyN when testing on a same CPU.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge