Jiangjie Wu
SUFFICIENT: A scan-specific unsupervised deep learning framework for high-resolution 3D isotropic fetal brain MRI reconstruction
May 26, 2025Abstract:High-quality 3D fetal brain MRI reconstruction from motion-corrupted 2D slices is crucial for clinical diagnosis. Reliable slice-to-volume registration (SVR)-based motion correction and super-resolution reconstruction (SRR) methods are essential. Deep learning (DL) has demonstrated potential in enhancing SVR and SRR when compared to conventional methods. However, it requires large-scale external training datasets, which are difficult to obtain for clinical fetal MRI. To address this issue, we propose an unsupervised iterative SVR-SRR framework for isotropic HR volume reconstruction. Specifically, SVR is formulated as a function mapping a 2D slice and a 3D target volume to a rigid transformation matrix, which aligns the slice to the underlying location in the target volume. The function is parameterized by a convolutional neural network, which is trained by minimizing the difference between the volume slicing at the predicted position and the input slice. In SRR, a decoding network embedded within a deep image prior framework is incorporated with a comprehensive image degradation model to produce the high-resolution (HR) volume. The deep image prior framework offers a local consistency prior to guide the reconstruction of HR volumes. By performing a forward degradation model, the HR volume is optimized by minimizing loss between predicted slices and the observed slices. Comprehensive experiments conducted on large-magnitude motion-corrupted simulation data and clinical data demonstrate the superior performance of the proposed framework over state-of-the-art fetal brain reconstruction frameworks.
RoCoSDF: Row-Column Scanned Neural Signed Distance Fields for Freehand 3D Ultrasound Imaging Shape Reconstruction
Aug 14, 2024



Abstract:The reconstruction of high-quality shape geometry is crucial for developing freehand 3D ultrasound imaging. However, the shape reconstruction of multi-view ultrasound data remains challenging due to the elevation distortion caused by thick transducer probes. In this paper, we present a novel learning-based framework RoCoSDF, which can effectively generate an implicit surface through continuous shape representations derived from row-column scanned datasets. In RoCoSDF, we encode the datasets from different views into the corresponding neural signed distance function (SDF) and then operate all SDFs in a normalized 3D space to restore the actual surface contour. Without requiring pre-training on large-scale ground truth shapes, our approach can synthesize a smooth and continuous signed distance field from multi-view SDFs to implicitly represent the actual geometry. Furthermore, two regularizers are introduced to facilitate shape refinement by constraining the SDF near the surface. The experiments on twelve shapes data acquired by two ultrasound transducer probes validate that RoCoSDF can effectively reconstruct accurate geometric shapes from multi-view ultrasound data, which outperforms current reconstruction methods. Code is available at https://github.com/chenhbo/RoCoSDF.
JSMoCo: Joint Coil Sensitivity and Motion Correction in Parallel MRI with a Self-Calibrating Score-Based Diffusion Model
Oct 14, 2023



Abstract:Magnetic Resonance Imaging (MRI) stands as a powerful modality in clinical diagnosis. However, it is known that MRI faces challenges such as long acquisition time and vulnerability to motion-induced artifacts. Despite the success of many existing motion correction algorithms, there has been limited research focused on correcting motion artifacts on the estimated coil sensitivity maps for fast MRI reconstruction. Existing methods might suffer from severe performance degradation due to error propagation resulting from the inaccurate coil sensitivity maps estimation. In this work, we propose to jointly estimate the motion parameters and coil sensitivity maps for under-sampled MRI reconstruction, referred to as JSMoCo. However, joint estimation of motion parameters and coil sensitivities results in a highly ill-posed inverse problem due to an increased number of unknowns. To address this, we introduce score-based diffusion models as powerful priors and leverage the MRI physical principles to efficiently constrain the solution space for this optimization problem. Specifically, we parameterize the rigid motion as three trainable variables and model coil sensitivity maps as polynomial functions. Leveraging the physical knowledge, we then employ Gibbs sampler for joint estimation, ensuring system consistency between sensitivity maps and desired images, avoiding error propagation from pre-estimated sensitivity maps to the reconstructed images. We conduct comprehensive experiments to evaluate the performance of JSMoCo on the fastMRI dataset. The results show that our method is capable of reconstructing high-quality MRI images from sparsely-sampled k-space data, even affected by motion. It achieves this by accurately estimating both motion parameters and coil sensitivities, effectively mitigating motion-related challenges during MRI reconstruction.
Self-supervised arbitrary scale super-resolution framework for anisotropic MRI
May 02, 2023Abstract:In this paper, we propose an efficient self-supervised arbitrary-scale super-resolution (SR) framework to reconstruct isotropic magnetic resonance (MR) images from anisotropic MRI inputs without involving external training data. The proposed framework builds a training dataset using in-the-wild anisotropic MR volumes with arbitrary image resolution. We then formulate the 3D volume SR task as a SR problem for 2D image slices. The anisotropic volume's high-resolution (HR) plane is used to build the HR-LR image pairs for model training. We further adapt the implicit neural representation (INR) network to implement the 2D arbitrary-scale image SR model. Finally, we leverage the well-trained proposed model to up-sample the 2D LR plane extracted from the anisotropic MR volumes to their HR views. The isotropic MR volumes thus can be reconstructed by stacking and averaging the generated HR slices. Our proposed framework has two major advantages: (1) It only involves the arbitrary-resolution anisotropic MR volumes, which greatly improves the model practicality in real MR imaging scenarios (e.g., clinical brain image acquisition); (2) The INR-based SR model enables arbitrary-scale image SR from the arbitrary-resolution input image, which significantly improves model training efficiency. We perform experiments on a simulated public adult brain dataset and a real collected 7T brain dataset. The results indicate that our current framework greatly outperforms two well-known self-supervised models for anisotropic MR image SR tasks.
Continuous longitudinal fetus brain atlas construction via implicit neural representation
Sep 14, 2022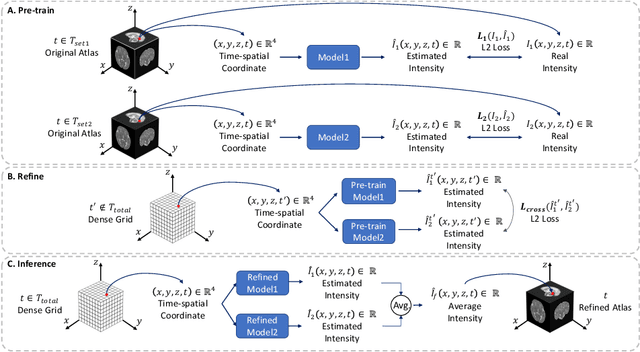
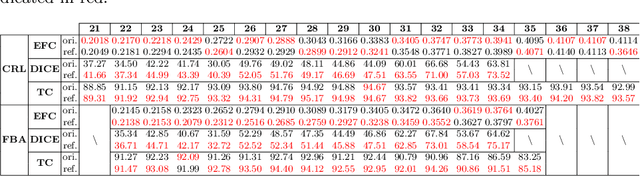
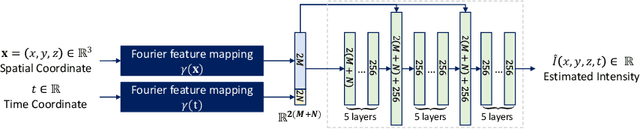
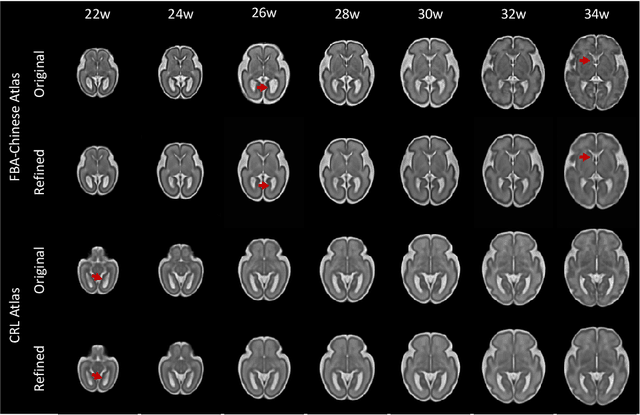
Abstract:Longitudinal fetal brain atlas is a powerful tool for understanding and characterizing the complex process of fetus brain development. Existing fetus brain atlases are typically constructed by averaged brain images on discrete time points independently over time. Due to the differences in onto-genetic trends among samples at different time points, the resulting atlases suffer from temporal inconsistency, which may lead to estimating error of the brain developmental characteristic parameters along the timeline. To this end, we proposed a multi-stage deep-learning framework to tackle the time inconsistency issue as a 4D (3D brain volume + 1D age) image data denoising task. Using implicit neural representation, we construct a continuous and noise-free longitudinal fetus brain atlas as a function of the 4D spatial-temporal coordinate. Experimental results on two public fetal brain atlases (CRL and FBA-Chinese atlases) show that the proposed method can significantly improve the atlas temporal consistency while maintaining good fetus brain structure representation. In addition, the continuous longitudinal fetus brain atlases can also be extensively applied to generate finer 4D atlases in both spatial and temporal resolution.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge