Xuanyu Tian
Improving 2D Diffusion Models for 3D Medical Imaging with Inter-Slice Consistent Stochasticity
Feb 04, 2026Abstract:3D medical imaging is in high demand and essential for clinical diagnosis and scientific research. Currently, diffusion models (DMs) have become an effective tool for medical imaging reconstruction thanks to their ability to learn rich, high-quality data priors. However, learning the 3D data distribution with DMs in medical imaging is challenging, not only due to the difficulties in data collection but also because of the significant computational burden during model training. A common compromise is to train the DMs on 2D data priors and reconstruct stacked 2D slices to address 3D medical inverse problems. However, the intrinsic randomness of diffusion sampling causes severe inter-slice discontinuities of reconstructed 3D volumes. Existing methods often enforce continuity regularizations along the z-axis, which introduces sensitive hyper-parameters and may lead to over-smoothing results. In this work, we revisit the origin of stochasticity in diffusion sampling and introduce Inter-Slice Consistent Stochasticity (ISCS), a simple yet effective strategy that encourages interslice consistency during diffusion sampling. Our key idea is to control the consistency of stochastic noise components during diffusion sampling, thereby aligning their sampling trajectories without adding any new loss terms or optimization steps. Importantly, the proposed ISCS is plug-and-play and can be dropped into any 2D trained diffusion based 3D reconstruction pipeline without additional computational cost. Experiments on several medical imaging problems show that our method can effectively improve the performance of medical 3D imaging problems based on 2D diffusion models. Our findings suggest that controlling inter-slice stochasticity is a principled and practically attractive route toward high-fidelity 3D medical imaging with 2D diffusion priors. The code is available at: https://github.com/duchenhe/ISCS
Unsupervised Motion-Compensated Decomposition for Cardiac MRI Reconstruction via Neural Representation
Nov 17, 2025Abstract:Cardiac magnetic resonance (CMR) imaging is widely used to characterize cardiac morphology and function. To accelerate CMR imaging, various methods have been proposed to recover high-quality spatiotemporal CMR images from highly undersampled k-t space data. However, current CMR reconstruction techniques either fail to achieve satisfactory image quality or are restricted by the scarcity of ground truth data, leading to limited applicability in clinical scenarios. In this work, we proposed MoCo-INR, a new unsupervised method that integrates implicit neural representations (INR) with the conventional motion-compensated (MoCo) framework. Using explicit motion modeling and the continuous prior of INRs, MoCo-INR can produce accurate cardiac motion decomposition and high-quality CMR reconstruction. Furthermore, we introduce a new INR network architecture tailored to the CMR problem, which significantly stabilizes model optimization. Experiments on retrospective (simulated) datasets demonstrate the superiority of MoCo-INR over state-of-the-art methods, achieving fast convergence and fine-detailed reconstructions at ultra-high acceleration factors (e.g., 20x in VISTA sampling). Additionally, evaluations on prospective (real-acquired) free-breathing CMR scans highlight the clinical practicality of MoCo-INR for real-time imaging. Several ablation studies further confirm the effectiveness of the critical components of MoCo-INR.
Unsupervised Self-Prior Embedding Neural Representation for Iterative Sparse-View CT Reconstruction
Feb 08, 2025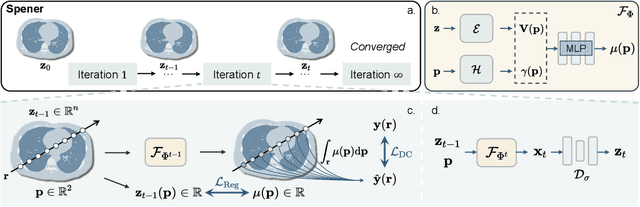
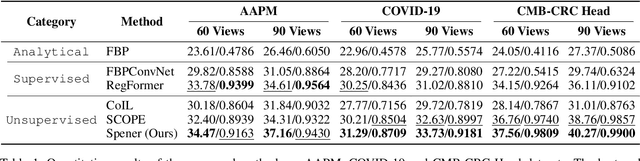
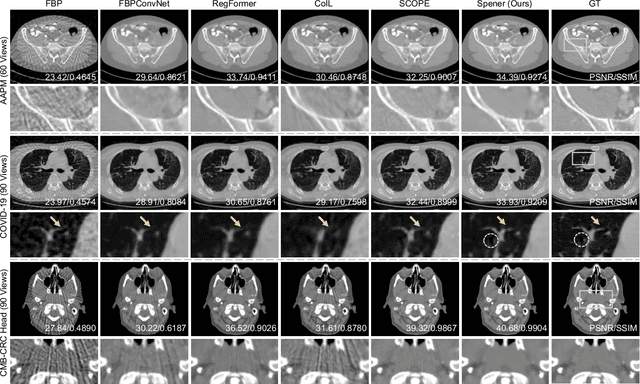
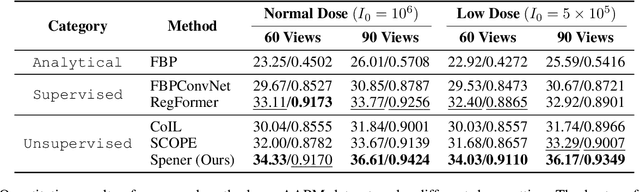
Abstract:Emerging unsupervised implicit neural representation (INR) methods, such as NeRP, NeAT, and SCOPE, have shown great potential to address sparse-view computed tomography (SVCT) inverse problems. Although these INR-based methods perform well in relatively dense SVCT reconstructions, they struggle to achieve comparable performance to supervised methods in sparser SVCT scenarios. They are prone to being affected by noise, limiting their applicability in real clinical settings. Additionally, current methods have not fully explored the use of image domain priors for solving SVCsT inverse problems. In this work, we demonstrate that imperfect reconstruction results can provide effective image domain priors for INRs to enhance performance. To leverage this, we introduce Self-prior embedding neural representation (Spener), a novel unsupervised method for SVCT reconstruction that integrates iterative reconstruction algorithms. During each iteration, Spener extracts local image prior features from the previous iteration and embeds them to constrain the solution space. Experimental results on multiple CT datasets show that our unsupervised Spener method achieves performance comparable to supervised state-of-the-art (SOTA) methods on in-domain data while outperforming them on out-of-domain datasets. Moreover, Spener significantly improves the performance of INR-based methods in handling SVCT with noisy sinograms. Our code is available at https://github.com/MeijiTian/Spener.
Zero-Shot Image Denoising for High-Resolution Electron Microscopy
Jun 20, 2024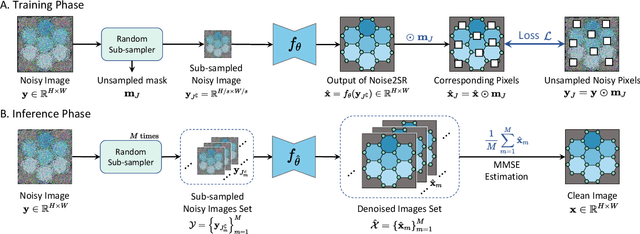
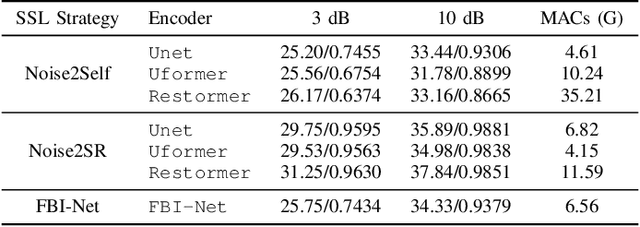
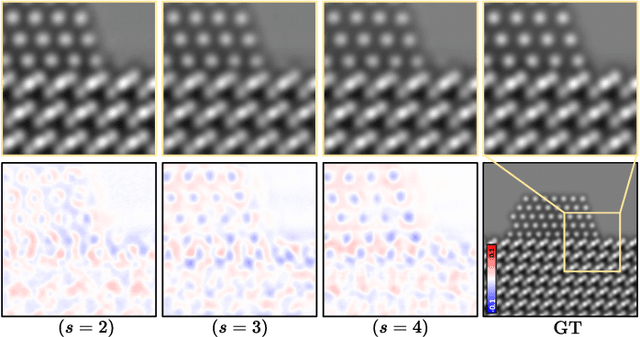
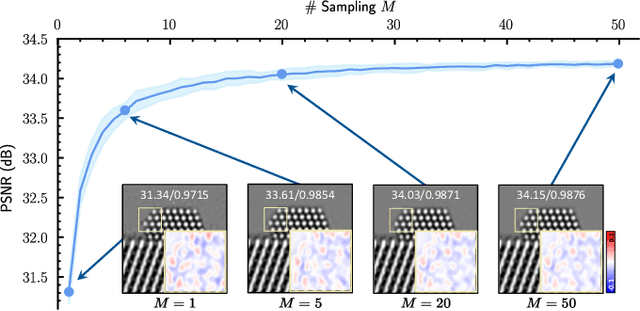
Abstract:High-resolution electron microscopy (HREM) imaging technique is a powerful tool for directly visualizing a broad range of materials in real-space. However, it faces challenges in denoising due to ultra-low signal-to-noise ratio (SNR) and scarce data availability. In this work, we propose Noise2SR, a zero-shot self-supervised learning (ZS-SSL) denoising framework for HREM. Within our framework, we propose a super-resolution (SR) based self-supervised training strategy, incorporating the Random Sub-sampler module. The Random Sub-sampler is designed to generate approximate infinite noisy pairs from a single noisy image, serving as an effective data augmentation in zero-shot denoising. Noise2SR trains the network with paired noisy images of different resolutions, which is conducted via SR strategy. The SR-based training facilitates the network adopting more pixels for supervision, and the random sub-sampling helps compel the network to learn continuous signals enhancing the robustness. Meanwhile, we mitigate the uncertainty caused by random-sampling by adopting minimum mean squared error (MMSE) estimation for the denoised results. With the distinctive integration of training strategy and proposed designs, Noise2SR can achieve superior denoising performance using a single noisy HREM image. We evaluate the performance of Noise2SR in both simulated and real HREM denoising tasks. It outperforms state-of-the-art ZS-SSL methods and achieves comparable denoising performance with supervised methods. The success of Noise2SR suggests its potential for improving the SNR of images in material imaging domains.
DPER: Diffusion Prior Driven Neural Representation for Limited Angle and Sparse View CT Reconstruction
Apr 27, 2024



Abstract:Limited-angle and sparse-view computed tomography (LACT and SVCT) are crucial for expanding the scope of X-ray CT applications. However, they face challenges due to incomplete data acquisition, resulting in diverse artifacts in the reconstructed CT images. Emerging implicit neural representation (INR) techniques, such as NeRF, NeAT, and NeRP, have shown promise in under-determined CT imaging reconstruction tasks. However, the unsupervised nature of INR architecture imposes limited constraints on the solution space, particularly for the highly ill-posed reconstruction task posed by LACT and ultra-SVCT. In this study, we introduce the Diffusion Prior Driven Neural Representation (DPER), an advanced unsupervised framework designed to address the exceptionally ill-posed CT reconstruction inverse problems. DPER adopts the Half Quadratic Splitting (HQS) algorithm to decompose the inverse problem into data fidelity and distribution prior sub-problems. The two sub-problems are respectively addressed by INR reconstruction scheme and pre-trained score-based diffusion model. This combination initially preserves the implicit image local consistency prior from INR. Additionally, it effectively augments the feasibility of the solution space for the inverse problem through the generative diffusion model, resulting in increased stability and precision in the solutions. We conduct comprehensive experiments to evaluate the performance of DPER on LACT and ultra-SVCT reconstruction with two public datasets (AAPM and LIDC). The results show that our method outperforms the state-of-the-art reconstruction methods on in-domain datasets, while achieving significant performance improvements on out-of-domain datasets.
JSMoCo: Joint Coil Sensitivity and Motion Correction in Parallel MRI with a Self-Calibrating Score-Based Diffusion Model
Oct 14, 2023



Abstract:Magnetic Resonance Imaging (MRI) stands as a powerful modality in clinical diagnosis. However, it is known that MRI faces challenges such as long acquisition time and vulnerability to motion-induced artifacts. Despite the success of many existing motion correction algorithms, there has been limited research focused on correcting motion artifacts on the estimated coil sensitivity maps for fast MRI reconstruction. Existing methods might suffer from severe performance degradation due to error propagation resulting from the inaccurate coil sensitivity maps estimation. In this work, we propose to jointly estimate the motion parameters and coil sensitivity maps for under-sampled MRI reconstruction, referred to as JSMoCo. However, joint estimation of motion parameters and coil sensitivities results in a highly ill-posed inverse problem due to an increased number of unknowns. To address this, we introduce score-based diffusion models as powerful priors and leverage the MRI physical principles to efficiently constrain the solution space for this optimization problem. Specifically, we parameterize the rigid motion as three trainable variables and model coil sensitivity maps as polynomial functions. Leveraging the physical knowledge, we then employ Gibbs sampler for joint estimation, ensuring system consistency between sensitivity maps and desired images, avoiding error propagation from pre-estimated sensitivity maps to the reconstructed images. We conduct comprehensive experiments to evaluate the performance of JSMoCo on the fastMRI dataset. The results show that our method is capable of reconstructing high-quality MRI images from sparsely-sampled k-space data, even affected by motion. It achieves this by accurately estimating both motion parameters and coil sensitivities, effectively mitigating motion-related challenges during MRI reconstruction.
Noise2SR: Learning to Denoise from Super-Resolved Single Noisy Fluorescence Image
Sep 14, 2022



Abstract:Fluorescence microscopy is a key driver to promote discoveries of biomedical research. However, with the limitation of microscope hardware and characteristics of the observed samples, the fluorescence microscopy images are susceptible to noise. Recently, a few self-supervised deep learning (DL) denoising methods have been proposed. However, the training efficiency and denoising performance of existing methods are relatively low in real scene noise removal. To address this issue, this paper proposed self-supervised image denoising method Noise2SR (N2SR) to train a simple and effective image denoising model based on single noisy observation. Our Noise2SR denoising model is designed for training with paired noisy images of different dimensions. Benefiting from this training strategy, Noise2SR is more efficiently self-supervised and able to restore more image details from a single noisy observation. Experimental results of simulated noise and real microscopy noise removal show that Noise2SR outperforms two blind-spot based self-supervised deep learning image denoising methods. We envision that Noise2SR has the potential to improve more other kind of scientific imaging quality.
* 12 pages, 6 figures
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge