Suchi Saria
Computer Science, Statistics, and Health Policy, Johns Hopkins University, Baltimore, MD, USA, ML, AI and Healthcare Lab, Bayesian Health, New York, NY, USA
Improving Coverage in Combined Prediction Sets with Weighted p-values
May 17, 2025Abstract:Conformal prediction quantifies the uncertainty of machine learning models by augmenting point predictions with valid prediction sets, assuming exchangeability. For complex scenarios involving multiple trials, models, or data sources, conformal prediction sets can be aggregated to create a prediction set that captures the overall uncertainty, often improving precision. However, aggregating multiple prediction sets with individual $1-\alpha$ coverage inevitably weakens the overall guarantee, typically resulting in $1-2\alpha$ worst-case coverage. In this work, we propose a framework for the weighted aggregation of prediction sets, where weights are assigned to each prediction set based on their contribution. Our framework offers flexible control over how the sets are aggregated, achieving tighter coverage bounds that interpolate between the $1-2\alpha$ guarantee of the combined models and the $1-\alpha$ guarantee of an individual model depending on the distribution of weights. We extend our framework to data-dependent weights, and we derive a general procedure for data-dependent weight aggregation that maintains finite-sample validity. We demonstrate the effectiveness of our methods through experiments on synthetic and real data in the mixture-of-experts setting, and we show that aggregation with data-dependent weights provides a form of adaptive coverage.
WATCH: Adaptive Monitoring for AI Deployments via Weighted-Conformal Martingales
May 12, 2025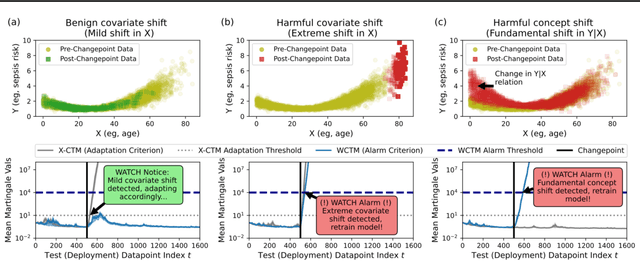
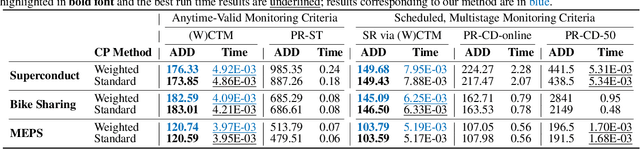
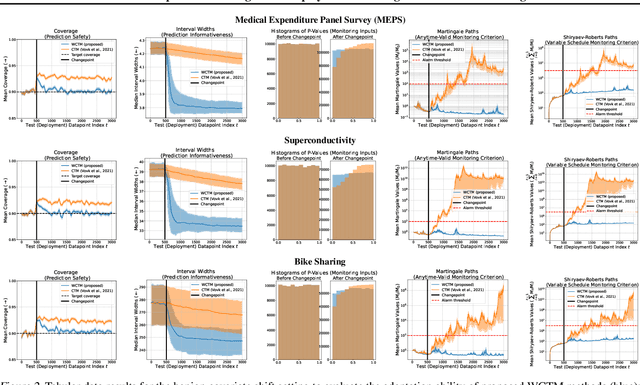
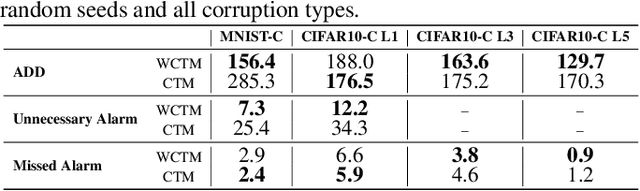
Abstract:Responsibly deploying artificial intelligence (AI) / machine learning (ML) systems in high-stakes settings arguably requires not only proof of system reliability, but moreover continual, post-deployment monitoring to quickly detect and address any unsafe behavior. Statistical methods for nonparametric change-point detection -- especially the tools of conformal test martingales (CTMs) and anytime-valid inference -- offer promising approaches to this monitoring task. However, existing methods are restricted to monitoring limited hypothesis classes or ``alarm criteria'' (such as data shifts that violate certain exchangeability assumptions), do not allow for online adaptation in response to shifts, and/or do not enable root-cause analysis of any degradation. In this paper, we expand the scope of these monitoring methods by proposing a weighted generalization of conformal test martingales (WCTMs), which lay a theoretical foundation for online monitoring for any unexpected changepoints in the data distribution while controlling false-alarms. For practical applications, we propose specific WCTM algorithms that adapt online to mild covariate shifts (in the marginal input distribution) while quickly detecting and diagnosing more severe shifts, such as concept shifts (in the conditional label distribution) or extreme (out-of-support) covariate shifts that cannot be easily adapted to. On real-world datasets, we demonstrate improved performance relative to state-of-the-art baselines.
WATCH: Weighted Adaptive Testing for Changepoint Hypotheses via Weighted-Conformal Martingales
May 07, 2025



Abstract:Responsibly deploying artificial intelligence (AI) / machine learning (ML) systems in high-stakes settings arguably requires not only proof of system reliability, but moreover continual, post-deployment monitoring to quickly detect and address any unsafe behavior. Statistical methods for nonparametric change-point detection -- especially the tools of conformal test martingales (CTMs) and anytime-valid inference -- offer promising approaches to this monitoring task. However, existing methods are restricted to monitoring limited hypothesis classes or ``alarm criteria,'' such as data shifts that violate certain exchangeability assumptions, or do not allow for online adaptation in response to shifts. In this paper, we expand the scope of these monitoring methods by proposing a weighted generalization of conformal test martingales (WCTMs), which lay a theoretical foundation for online monitoring for any unexpected changepoints in the data distribution while controlling false-alarms. For practical applications, we propose specific WCTM algorithms that accommodate online adaptation to mild covariate shifts (in the marginal input distribution) while raising alarms in response to more severe shifts, such as concept shifts (in the conditional label distribution) or extreme (out-of-support) covariate shifts that cannot be easily adapted to. On real-world datasets, we demonstrate improved performance relative to state-of-the-art baselines.
Between Linear and Sinusoidal: Rethinking the Time Encoder in Dynamic Graph Learning
Apr 10, 2025
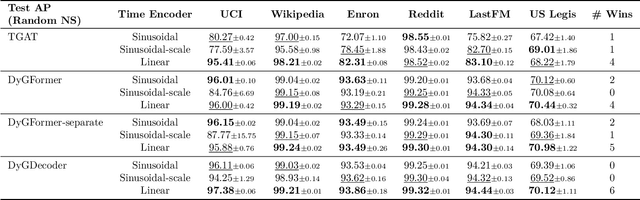
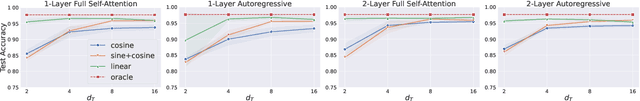

Abstract:Dynamic graph learning is essential for applications involving temporal networks and requires effective modeling of temporal relationships. Seminal attention-based models like TGAT and DyGFormer rely on sinusoidal time encoders to capture temporal relationships between edge events. In this paper, we study a simpler alternative: the linear time encoder, which avoids temporal information loss caused by sinusoidal functions and reduces the need for high dimensional time encoders. We show that the self-attention mechanism can effectively learn to compute time spans from linear time encodings and extract relevant temporal patterns. Through extensive experiments on six dynamic graph datasets, we demonstrate that the linear time encoder improves the performance of TGAT and DyGFormer in most cases. Moreover, the linear time encoder can lead to significant savings in model parameters with minimal performance loss. For example, compared to a 100-dimensional sinusoidal time encoder, TGAT with a 2-dimensional linear time encoder saves 43% of parameters and achieves higher average precision on five datasets. These results can be readily used to positively impact the design choices of a wide variety of dynamic graph learning architectures. The experimental code is available at: https://github.com/hsinghuan/dg-linear-time.git.
On Expert Estimation in Hierarchical Mixture of Experts: Beyond Softmax Gating Functions
Oct 03, 2024



Abstract:With the growing prominence of the Mixture of Experts (MoE) architecture in developing large-scale foundation models, we investigate the Hierarchical Mixture of Experts (HMoE), a specialized variant of MoE that excels in handling complex inputs and improving performance on targeted tasks. Our investigation highlights the advantages of using varied gating functions, moving beyond softmax gating within HMoE frameworks. We theoretically demonstrate that applying tailored gating functions to each expert group allows HMoE to achieve robust results, even when optimal gating functions are applied only at select hierarchical levels. Empirical validation across diverse scenarios supports these theoretical claims. This includes large-scale multimodal tasks, image classification, and latent domain discovery and prediction tasks, where our modified HMoE models show great performance improvements.
Conformal Validity Guarantees Exist for Any Data Distribution
May 10, 2024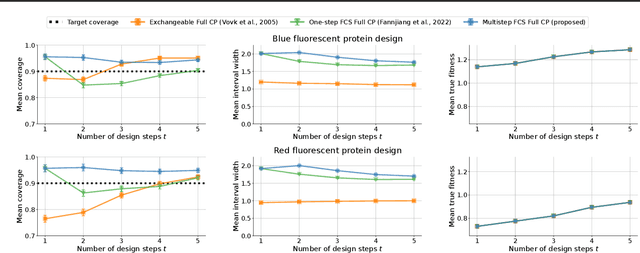
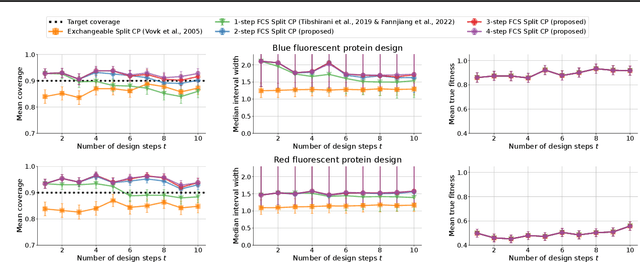
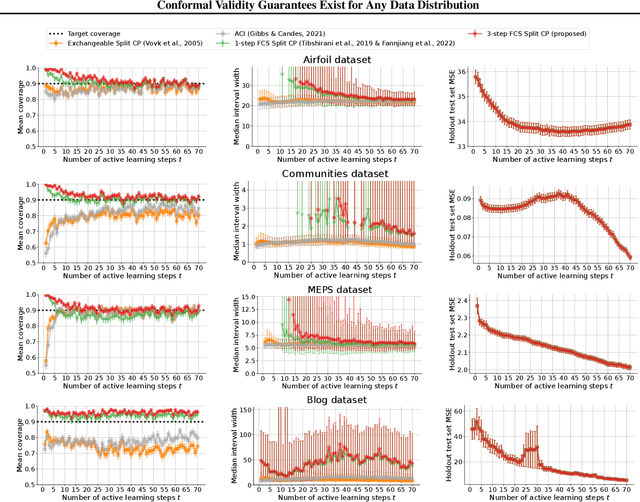
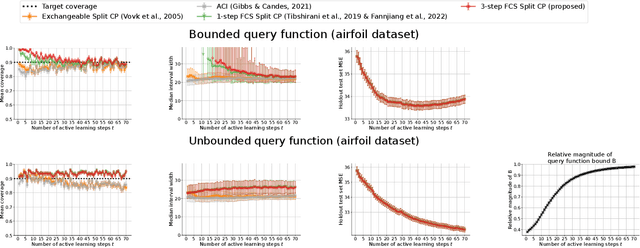
Abstract:As machine learning (ML) gains widespread adoption, practitioners are increasingly seeking means to quantify and control the risk these systems incur. This challenge is especially salient when ML systems have autonomy to collect their own data, such as in black-box optimization and active learning, where their actions induce sequential feedback-loop shifts in the data distribution. Conformal prediction has emerged as a promising approach to uncertainty and risk quantification, but existing variants either fail to accommodate sequences of data-dependent shifts, or do not fully exploit the fact that agent-induced shift is under our control. In this work we prove that conformal prediction can theoretically be extended to \textit{any} joint data distribution, not just exchangeable or quasi-exchangeable ones, although it is exceedingly impractical to compute in the most general case. For practical applications, we outline a procedure for deriving specific conformal algorithms for any data distribution, and we use this procedure to derive tractable algorithms for a series of agent-induced covariate shifts. We evaluate the proposed algorithms empirically on synthetic black-box optimization and active learning tasks.
FuseMoE: Mixture-of-Experts Transformers for Fleximodal Fusion
Feb 05, 2024



Abstract:As machine learning models in critical fields increasingly grapple with multimodal data, they face the dual challenges of handling a wide array of modalities, often incomplete due to missing elements, and the temporal irregularity and sparsity of collected samples. Successfully leveraging this complex data, while overcoming the scarcity of high-quality training samples, is key to improving these models' predictive performance. We introduce ``FuseMoE'', a mixture-of-experts framework incorporated with an innovative gating function. Designed to integrate a diverse number of modalities, FuseMoE is effective in managing scenarios with missing modalities and irregularly sampled data trajectories. Theoretically, our unique gating function contributes to enhanced convergence rates, leading to better performance in multiple downstream tasks. The practical utility of FuseMoE in real world is validated by a challenging set of clinical risk prediction tasks.
Causal-structure Driven Augmentations for Text OOD Generalization
Oct 19, 2023



Abstract:The reliance of text classifiers on spurious correlations can lead to poor generalization at deployment, raising concerns about their use in safety-critical domains such as healthcare. In this work, we propose to use counterfactual data augmentation, guided by knowledge of the causal structure of the data, to simulate interventions on spurious features and to learn more robust text classifiers. We show that this strategy is appropriate in prediction problems where the label is spuriously correlated with an attribute. Under the assumptions of such problems, we discuss the favorable sample complexity of counterfactual data augmentation, compared to importance re-weighting. Pragmatically, we match examples using auxiliary data, based on diff-in-diff methodology, and use a large language model (LLM) to represent a conditional probability of text. Through extensive experimentation on learning caregiver-invariant predictors of clinical diagnoses from medical narratives and on semi-synthetic data, we demonstrate that our method for simulating interventions improves out-of-distribution (OOD) accuracy compared to baseline invariant learning algorithms.
JAWS: Predictive Inference Under Covariate Shift
Jul 21, 2022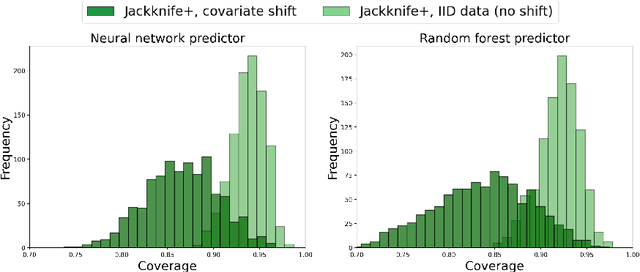
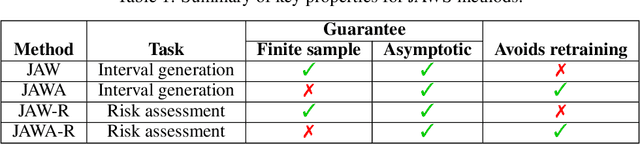
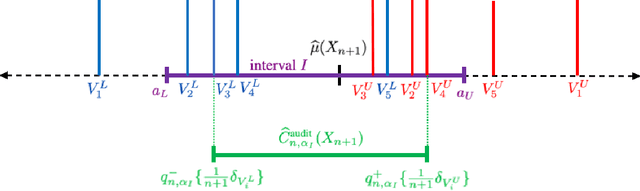
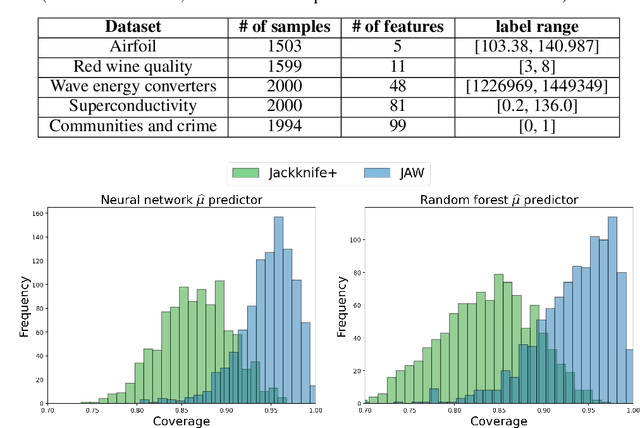
Abstract:We propose \textbf{JAWS}, a series of wrapper methods for distribution-free uncertainty quantification tasks under covariate shift, centered on our core method \textbf{JAW}, the \textbf{JA}ckknife+ \textbf{W}eighted with likelihood-ratio weights. JAWS also includes computationally efficient \textbf{A}pproximations of JAW using higher-order influence functions: \textbf{JAWA}. Theoretically, we show that JAW relaxes the jackknife+'s assumption of data exchangeability to achieve the same finite-sample coverage guarantee even under covariate shift. JAWA further approaches the JAW guarantee in the limit of either the sample size or the influence function order under mild assumptions. Moreover, we propose a general approach to repurposing any distribution-free uncertainty quantification method and its guarantees to the task of risk assessment: a task that generates the estimated probability that the true label lies within a user-specified interval. We then propose \textbf{JAW-R} and \textbf{JAWA-R} as the repurposed versions of proposed methods for \textbf{R}isk assessment. Practically, JAWS outperform the state-of-the-art predictive inference baselines in a variety of biased real world data sets for both interval-generation and risk-assessment auditing tasks.
Beyond Low Earth Orbit: Biomonitoring, Artificial Intelligence, and Precision Space Health
Dec 22, 2021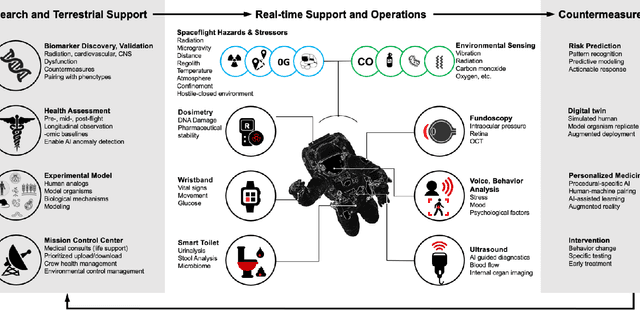
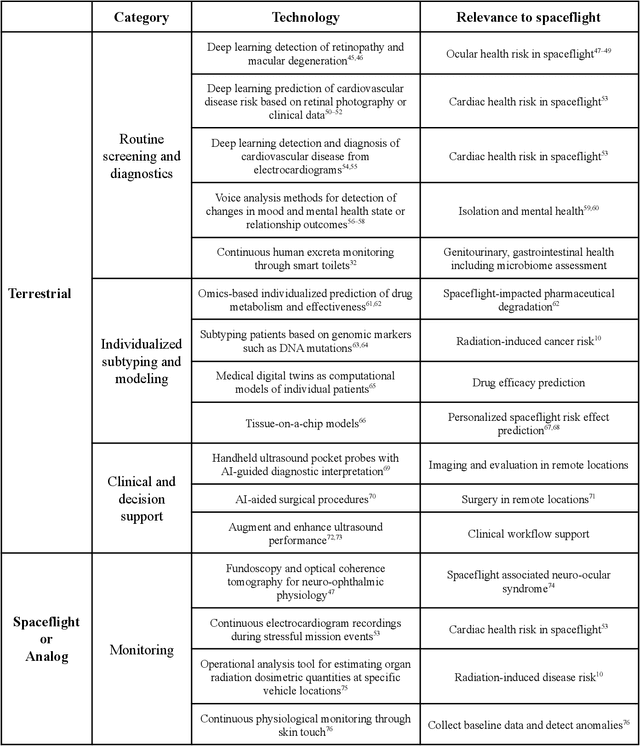
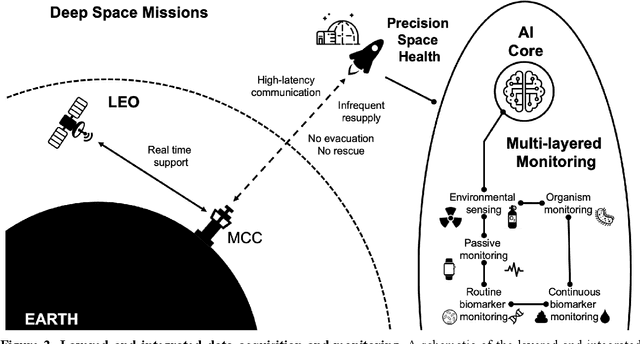
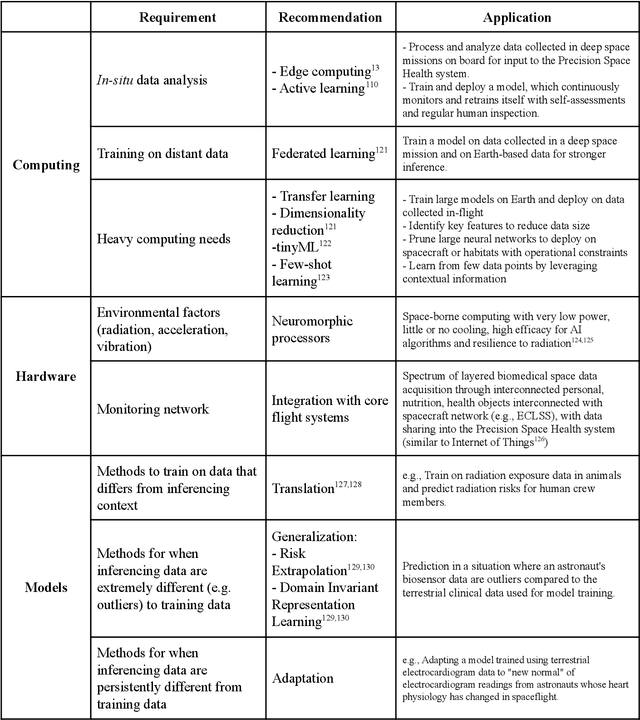
Abstract:Human space exploration beyond low Earth orbit will involve missions of significant distance and duration. To effectively mitigate myriad space health hazards, paradigm shifts in data and space health systems are necessary to enable Earth-independence, rather than Earth-reliance. Promising developments in the fields of artificial intelligence and machine learning for biology and health can address these needs. We propose an appropriately autonomous and intelligent Precision Space Health system that will monitor, aggregate, and assess biomedical statuses; analyze and predict personalized adverse health outcomes; adapt and respond to newly accumulated data; and provide preventive, actionable, and timely insights to individual deep space crew members and iterative decision support to their crew medical officer. Here we present a summary of recommendations from a workshop organized by the National Aeronautics and Space Administration, on future applications of artificial intelligence in space biology and health. In the next decade, biomonitoring technology, biomarker science, spacecraft hardware, intelligent software, and streamlined data management must mature and be woven together into a Precision Space Health system to enable humanity to thrive in deep space.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge