Shengjia Chen
Predict Patient Self-reported Race from Skin Histological Images
Jul 30, 2025Abstract:Artificial Intelligence (AI) has demonstrated success in computational pathology (CPath) for disease detection, biomarker classification, and prognosis prediction. However, its potential to learn unintended demographic biases, particularly those related to social determinants of health, remains understudied. This study investigates whether deep learning models can predict self-reported race from digitized dermatopathology slides and identifies potential morphological shortcuts. Using a multisite dataset with a racially diverse population, we apply an attention-based mechanism to uncover race-associated morphological features. After evaluating three dataset curation strategies to control for confounding factors, the final experiment showed that White and Black demographic groups retained high prediction performance (AUC: 0.799, 0.762), while overall performance dropped to 0.663. Attention analysis revealed the epidermis as a key predictive feature, with significant performance declines when these regions were removed. These findings highlight the need for careful data curation and bias mitigation to ensure equitable AI deployment in pathology. Code available at: https://github.com/sinai-computational-pathology/CPath_SAIF.
Weakly-supervised Contrastive Learning with Quantity Prompts for Moving Infrared Small Target Detection
Jul 03, 2025Abstract:Different from general object detection, moving infrared small target detection faces huge challenges due to tiny target size and weak background contrast.Currently, most existing methods are fully-supervised, heavily relying on a large number of manual target-wise annotations. However, manually annotating video sequences is often expensive and time-consuming, especially for low-quality infrared frame images. Inspired by general object detection, non-fully supervised strategies ($e.g.$, weakly supervised) are believed to be potential in reducing annotation requirements. To break through traditional fully-supervised frameworks, as the first exploration work, this paper proposes a new weakly-supervised contrastive learning (WeCoL) scheme, only requires simple target quantity prompts during model training.Specifically, in our scheme, based on the pretrained segment anything model (SAM), a potential target mining strategy is designed to integrate target activation maps and multi-frame energy accumulation.Besides, contrastive learning is adopted to further improve the reliability of pseudo-labels, by calculating the similarity between positive and negative samples in feature subspace.Moreover, we propose a long-short term motion-aware learning scheme to simultaneously model the local motion patterns and global motion trajectory of small targets.The extensive experiments on two public datasets (DAUB and ITSDT-15K) verify that our weakly-supervised scheme could often outperform early fully-supervised methods. Even, its performance could reach over 90\% of state-of-the-art (SOTA) fully-supervised ones.
A Clinical Benchmark of Public Self-Supervised Pathology Foundation Models
Jul 11, 2024



Abstract:The use of self-supervised learning (SSL) to train pathology foundation models has increased substantially in the past few years. Notably, several models trained on large quantities of clinical data have been made publicly available in recent months. This will significantly enhance scientific research in computational pathology and help bridge the gap between research and clinical deployment. With the increase in availability of public foundation models of different sizes, trained using different algorithms on different datasets, it becomes important to establish a benchmark to compare the performance of such models on a variety of clinically relevant tasks spanning multiple organs and diseases. In this work, we present a collection of pathology datasets comprising clinical slides associated with clinically relevant endpoints including cancer diagnoses and a variety of biomarkers generated during standard hospital operation from two medical centers. We leverage these datasets to systematically assess the performance of public pathology foundation models and provide insights into best practices for training new foundation models and selecting appropriate pretrained models.
Benchmarking Embedding Aggregation Methods in Computational Pathology: A Clinical Data Perspective
Jul 10, 2024


Abstract:Recent advances in artificial intelligence (AI), in particular self-supervised learning of foundation models (FMs), are revolutionizing medical imaging and computational pathology (CPath). A constant challenge in the analysis of digital Whole Slide Images (WSIs) is the problem of aggregating tens of thousands of tile-level image embeddings to a slide-level representation. Due to the prevalent use of datasets created for genomic research, such as TCGA, for method development, the performance of these techniques on diagnostic slides from clinical practice has been inadequately explored. This study conducts a thorough benchmarking analysis of ten slide-level aggregation techniques across nine clinically relevant tasks, including diagnostic assessment, biomarker classification, and outcome prediction. The results yield following key insights: (1) Embeddings derived from domain-specific (histological images) FMs outperform those from generic ImageNet-based models across aggregation methods. (2) Spatial-aware aggregators enhance the performance significantly when using ImageNet pre-trained models but not when using FMs. (3) No single model excels in all tasks and spatially-aware models do not show general superiority as it would be expected. These findings underscore the need for more adaptable and universally applicable aggregation techniques, guiding future research towards tools that better meet the evolving needs of clinical-AI in pathology. The code used in this work is available at \url{https://github.com/fuchs-lab-public/CPath_SABenchmark}.
Triple-domain Feature Learning with Frequency-aware Memory Enhancement for Moving Infrared Small Target Detection
Jun 11, 2024Abstract:Moving infrared small target detection presents significant challenges due to tiny target sizes and low contrast against backgrounds. Currently-existing methods primarily focus on extracting target features only from the spatial-temporal domain. For further enhancing feature representation, more information domains such as frequency are believed to be potentially valuable. To extend target feature learning, we propose a new Triple-domain Strategy (Tridos) with the frequency-aware memory enhancement on the spatial-temporal domain. In our scheme, it effectively detaches and enhances frequency features by a local-global frequency-aware module with Fourier transform. Inspired by the human visual system, our memory enhancement aims to capture the target spatial relations between video frames. Furthermore, it encodes temporal dynamics motion features via differential learning and residual enhancing. Additionally, we further design a residual compensation unit to reconcile possible cross-domain feature mismatches. To our best knowledge, our Tridos is the first work to explore target feature learning comprehensively in spatial-temporal-frequency domains. The extensive experiments on three datasets (DAUB, ITSDT-15K, and IRDST) validate that our triple-domain learning scheme could be obviously superior to state-of-the-art ones. Source codes are available at https://github.com/UESTC-nnLab/Tridos.
MR-Transformer: Vision Transformer for Total Knee Replacement Prediction Using Magnetic Resonance Imaging
May 05, 2024Abstract:A transformer-based deep learning model, MR-Transformer, was developed for total knee replacement (TKR) prediction using magnetic resonance imaging (MRI). The model incorporates the ImageNet pre-training and captures three-dimensional (3D) spatial correlation from the MR images. The performance of the proposed model was compared to existing state-of-the-art deep learning models for knee injury diagnosis using MRI. Knee MR scans of four different tissue contrasts from the Osteoarthritis Initiative and Multicenter Osteoarthritis Study databases were utilized in the study. Experimental results demonstrated the state-of-the-art performance of the proposed model on TKR prediction using MRI.
Estimation of Time-to-Total Knee Replacement Surgery
Apr 29, 2024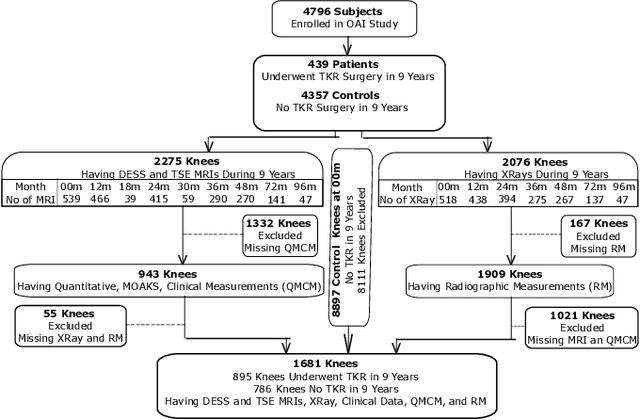
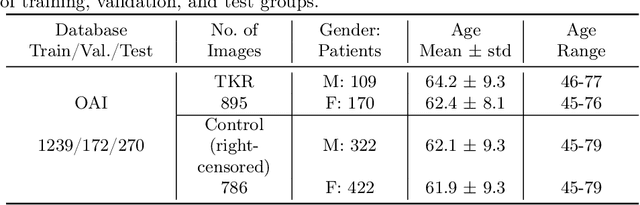
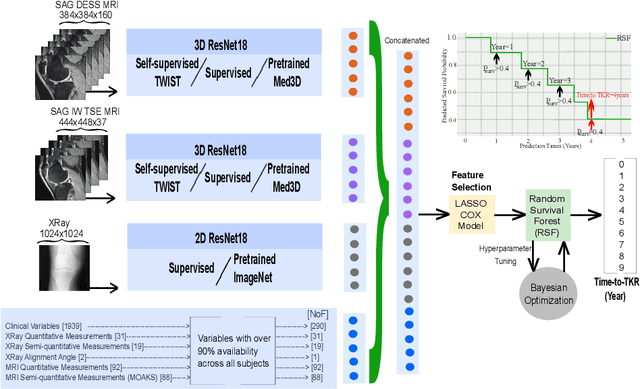
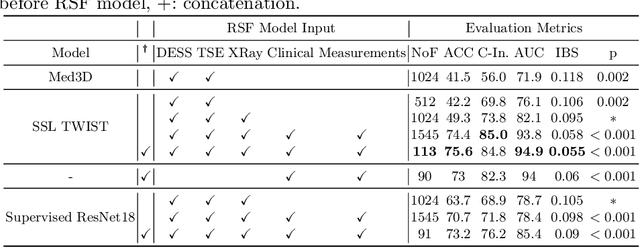
Abstract:A survival analysis model for predicting time-to-total knee replacement (TKR) was developed using features from medical images and clinical measurements. Supervised and self-supervised deep learning approaches were utilized to extract features from radiographs and magnetic resonance images. Extracted features were combined with clinical and image assessments for survival analysis using random survival forests. The proposed model demonstrated high discrimination power by combining deep learning features and clinical and image assessments using a fusion of multiple modalities. The model achieved an accuracy of 75.6% and a C-Index of 84.8% for predicting the time-to-TKR surgery. Accurate time-to-TKR predictions have the potential to help assist physicians to personalize treatment strategies and improve patient outcomes.
Prediction of drug effectiveness in rheumatoid arthritis patients based on machine learning algorithms
Oct 18, 2022



Abstract:Rheumatoid arthritis (RA) is an autoimmune condition caused when patients' immune system mistakenly targets their own tissue. Machine learning (ML) has the potential to identify patterns in patient electronic health records (EHR) to forecast the best clinical treatment to improve patient outcomes. This study introduced a Drug Response Prediction (DRP) framework with two main goals: 1) design a data processing pipeline to extract information from tabular clinical data, and then preprocess it for functional use, and 2) predict RA patient's responses to drugs and evaluate classification models' performance. We propose a novel two-stage ML framework based on European Alliance of Associations for Rheumatology (EULAR) criteria cutoffs to model drug effectiveness. Our model Stacked-Ensemble DRP was developed and cross-validated using data from 425 RA patients. The evaluation used a subset of 124 patients (30%) from the same data source. In the evaluation of the test set, two-stage DRP leads to improved classification accuracy over other end-to-end classification models for binary classification. Our proposed method provides a complete pipeline to predict disease activity scores and identify the group that does not respond well to anti-TNF treatments, thus showing promise in supporting clinical decisions based on EHR information.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge