Daniel K. Sodickson
A Trust-Guided Approach to MR Image Reconstruction with Side Information
Jan 06, 2025



Abstract:Reducing MRI scan times can improve patient care and lower healthcare costs. Many acceleration methods are designed to reconstruct diagnostic-quality images from limited sets of acquired $\textit{k}$-space data. This task can be framed as a linear inverse problem (LIP), where, as a result of undersampling, the forward operator may become rank-deficient or exhibit small singular values. This results in ambiguities in reconstruction, in which multiple generally incorrect or non-diagnostic images can map to the same acquired data. To address such ambiguities, it is crucial to incorporate prior knowledge, for example in the form of regularization. Another form of prior knowledge less commonly used in medical imaging is contextual side information garnered from other sources than the current acquisition. Here, we propose the $\textbf{T}$rust-$\textbf{G}$uided $\textbf{V}$ariational $\textbf{N}$etwork $\textbf{(TGVN)}$, a novel end-to-end deep learning framework that effectively integrates side information into LIPs. TGVN eliminates undesirable solutions from the ambiguous space of the forward operator while remaining faithful to the acquired data. We demonstrate its effectiveness in multi-coil, multi-contrast MR image reconstruction, where incomplete or low-quality measurements from one contrast are used as side information to reconstruct high-quality images of another contrast from heavily under-sampled data. Our method is robust across different contrasts, anatomies, and field strengths. Compared to baselines that also utilize side information, TGVN achieves superior image quality at challenging under-sampling levels, drastically speeding up acquisition while minimizing hallucinations. Our approach is also versatile enough to incorporate many different types of side information (including previous scans or even text) into any LIP.
An experimental system for detection and localization of hemorrhage using ultra-wideband microwaves with deep learning
Oct 03, 2023Abstract:Stroke is a leading cause of mortality and disability. Emergent diagnosis and intervention are critical, and predicated upon initial brain imaging; however, existing clinical imaging modalities are generally costly, immobile, and demand highly specialized operation and interpretation. Low-energy microwaves have been explored as low-cost, small form factor, fast, and safe probes of tissue dielectric properties, with both imaging and diagnostic potential. Nevertheless, challenges inherent to microwave reconstruction have impeded progress, hence microwave imaging (MWI) remains an elusive scientific aim. Herein, we introduce a dedicated experimental framework comprising a robotic navigation system to translate blood-mimicking phantoms within an anatomically realistic human head model. An 8-element ultra-wideband (UWB) array of modified antipodal Vivaldi antennas was developed and driven by a two-port vector network analyzer spanning 0.6-9.0 GHz at an operating power of 1 mw. Complex scattering parameters were measured, and dielectric signatures of hemorrhage were learned using a dedicated deep neural network for prediction of hemorrhage classes and localization. An overall sensitivity and specificity for detection >0.99 was observed, with Rayliegh mean localization error of 1.65 mm. The study establishes the feasibility of a robust experimental model and deep learning solution for UWB microwave stroke detection.
On the Feasibility of Machine Learning Augmented Magnetic Resonance for Point-of-Care Identification of Disease
Feb 02, 2023



Abstract:Early detection of many life-threatening diseases (e.g., prostate and breast cancer) within at-risk population can improve clinical outcomes and reduce cost of care. While numerous disease-specific "screening" tests that are closer to Point-of-Care (POC) are in use for this task, their low specificity results in unnecessary biopsies, leading to avoidable patient trauma and wasteful healthcare spending. On the other hand, despite the high accuracy of Magnetic Resonance (MR) imaging in disease diagnosis, it is not used as a POC disease identification tool because of poor accessibility. The root cause of poor accessibility of MR stems from the requirement to reconstruct high-fidelity images, as it necessitates a lengthy and complex process of acquiring large quantities of high-quality k-space measurements. In this study we explore the feasibility of an ML-augmented MR pipeline that directly infers the disease sidestepping the image reconstruction process. We hypothesise that the disease classification task can be solved using a very small tailored subset of k-space data, compared to image reconstruction. Towards that end, we propose a method that performs two tasks: 1) identifies a subset of the k-space that maximizes disease identification accuracy, and 2) infers the disease directly using the identified k-space subset, bypassing the image reconstruction step. We validate our hypothesis by measuring the performance of the proposed system across multiple diseases and anatomies. We show that comparable performance to image-based classifiers, trained on images reconstructed with full k-space data, can be achieved using small quantities of data: 8% of the data for detecting multiple abnormalities in prostate and brain scans, and 5% of the data for knee abnormalities. To better understand the proposed approach and instigate future research, we provide an extensive analysis and release code.
Differences between human and machine perception in medical diagnosis
Nov 28, 2020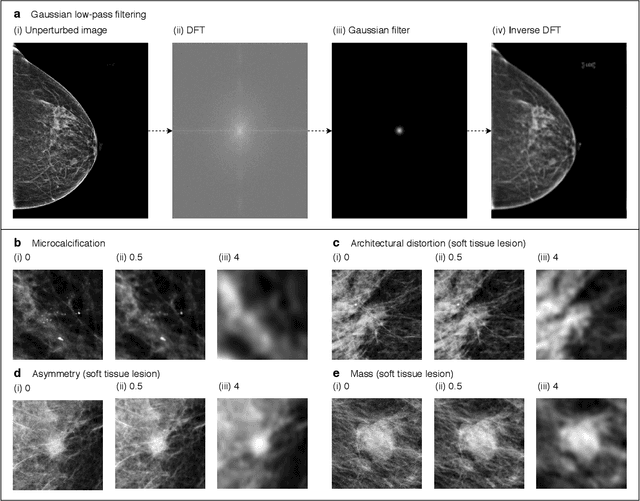
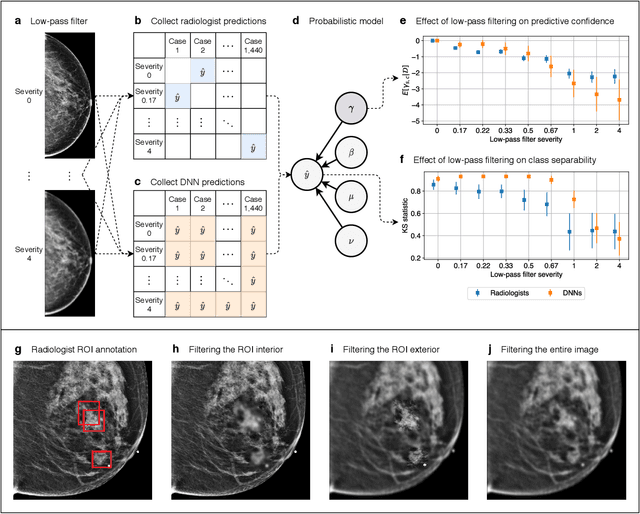
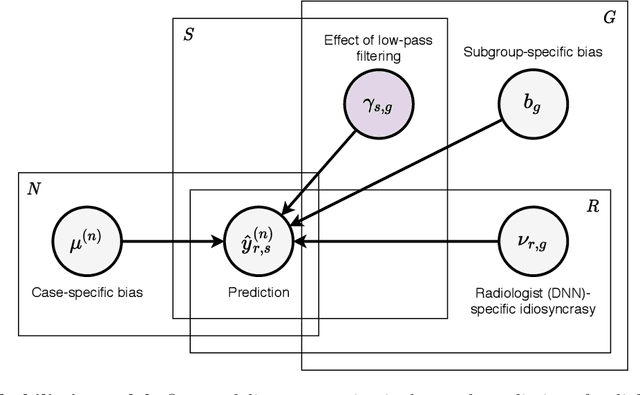
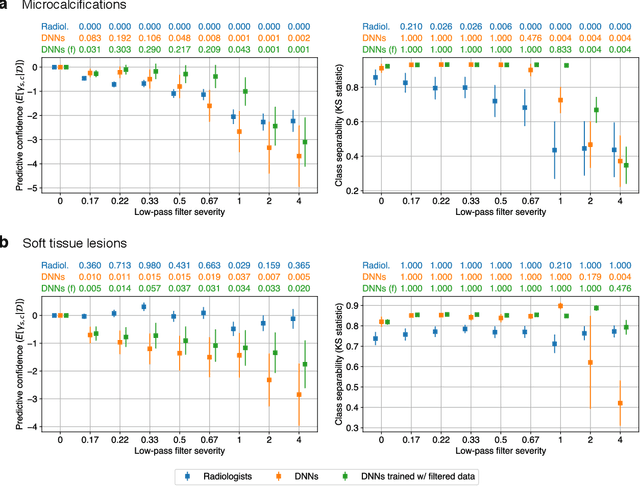
Abstract:Deep neural networks (DNNs) show promise in image-based medical diagnosis, but cannot be fully trusted since their performance can be severely degraded by dataset shifts to which human perception remains invariant. If we can better understand the differences between human and machine perception, we can potentially characterize and mitigate this effect. We therefore propose a framework for comparing human and machine perception in medical diagnosis. The two are compared with respect to their sensitivity to the removal of clinically meaningful information, and to the regions of an image deemed most suspicious. Drawing inspiration from the natural image domain, we frame both comparisons in terms of perturbation robustness. The novelty of our framework is that separate analyses are performed for subgroups with clinically meaningful differences. We argue that this is necessary in order to avert Simpson's paradox and draw correct conclusions. We demonstrate our framework with a case study in breast cancer screening, and reveal significant differences between radiologists and DNNs. We compare the two with respect to their robustness to Gaussian low-pass filtering, performing a subgroup analysis on microcalcifications and soft tissue lesions. For microcalcifications, DNNs use a separate set of high frequency components than radiologists, some of which lie outside the image regions considered most suspicious by radiologists. These features run the risk of being spurious, but if not, could represent potential new biomarkers. For soft tissue lesions, the divergence between radiologists and DNNs is even starker, with DNNs relying heavily on spurious high frequency components ignored by radiologists. Importantly, this deviation in soft tissue lesions was only observable through subgroup analysis, which highlights the importance of incorporating medical domain knowledge into our comparison framework.
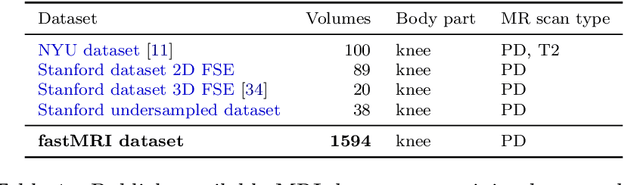
Advancing machine learning for MR image reconstruction with an open competition: Overview of the 2019 fastMRI challenge
Jan 06, 2020
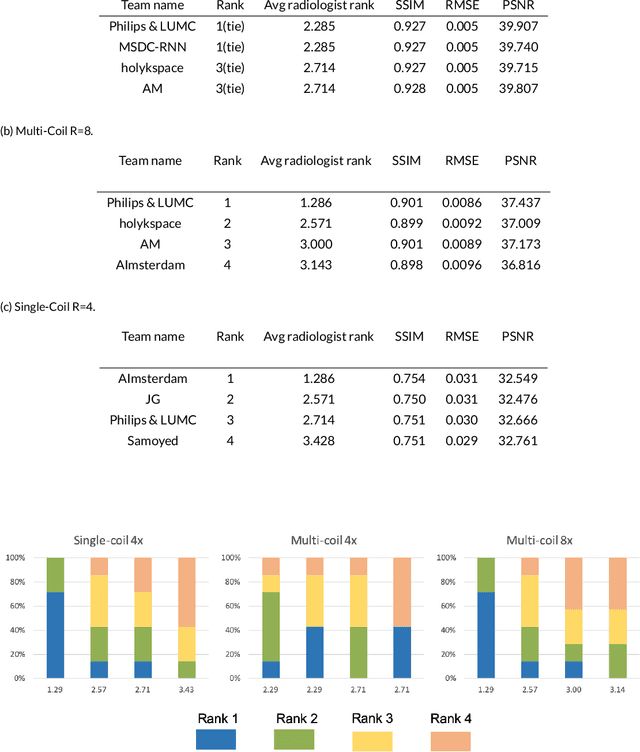
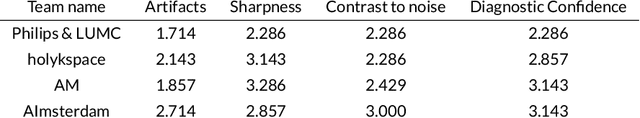
Abstract:Purpose: To advance research in the field of machine learning for MR image reconstruction with an open challenge. Methods: We provided participants with a dataset of raw k-space data from 1,594 consecutive clinical exams of the knee. The goal of the challenge was to reconstruct images from these data. In order to strike a balance between realistic data and a shallow learning curve for those not already familiar with MR image reconstruction, we ran multiple tracks for multi-coil and single-coil data. We performed a two-stage evaluation based on quantitative image metrics followed by evaluation by a panel of radiologists. The challenge ran from June to December of 2019. Results: We received a total of 33 challenge submissions. All participants chose to submit results from supervised machine learning approaches. Conclusion: The challenge led to new developments in machine learning for image reconstruction, provided insight into the current state of the art in the field, and highlighted remaining hurdles for clinical adoption.
GrappaNet: Combining Parallel Imaging with Deep Learning for Multi-Coil MRI Reconstruction
Nov 04, 2019

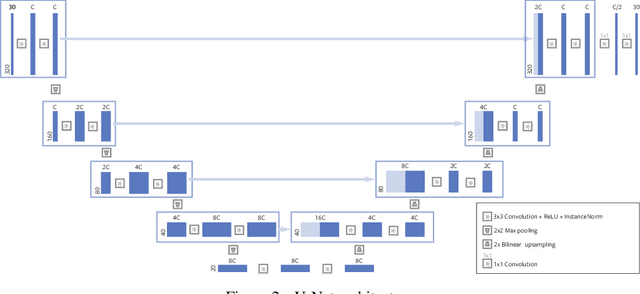
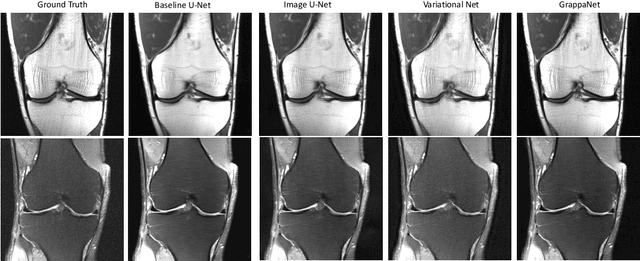
Abstract:Magnetic Resonance Image (MRI) acquisition is an inherently slow process which has spurred the development of two different acceleration methods: acquiring multiple correlated samples simultaneously (parallel imaging) and acquiring fewer samples than necessary for traditional signal processing methods (compressed sensing). Both methods provide complementary approaches to accelerating the speed of MRI acquisition. In this paper, we present a novel method to integrate traditional parallel imaging methods into deep neural networks that is able to generate high quality reconstructions even for high acceleration factors. The proposed method, called GrappaNet, performs progressive reconstruction by first mapping the reconstruction problem to a simpler one that can be solved by a traditional parallel imaging methods using a neural network, followed by an application of a parallel imaging method, and finally fine-tuning the output with another neural network. The entire network can be trained end-to-end. We present experimental results on the recently released fastMRI dataset and show that GrappaNet can generate higher quality reconstructions than competing methods for both $4\times$ and $8\times$ acceleration.
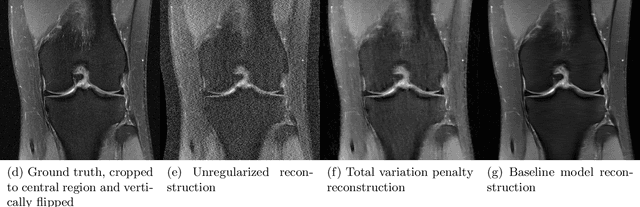
Deep Learning Methods for Parallel Magnetic Resonance Image Reconstruction
Apr 01, 2019



Abstract:Following the success of deep learning in a wide range of applications, neural network-based machine learning techniques have received interest as a means of accelerating magnetic resonance imaging (MRI). A number of ideas inspired by deep learning techniques from computer vision and image processing have been successfully applied to non-linear image reconstruction in the spirit of compressed sensing for both low dose computed tomography and accelerated MRI. The additional integration of multi-coil information to recover missing k-space lines in the MRI reconstruction process, is still studied less frequently, even though it is the de-facto standard for currently used accelerated MR acquisitions. This manuscript provides an overview of the recent machine learning approaches that have been proposed specifically for improving parallel imaging. A general background introduction to parallel MRI is given that is structured around the classical view of image space and k-space based methods. Both linear and non-linear methods are covered, followed by a discussion of recent efforts to further improve parallel imaging using machine learning, and specifically using artificial neural networks. Image-domain based techniques that introduce improved regularizers are covered as well as k-space based methods, where the focus is on better interpolation strategies using neural networks. Issues and open problems are discussed as well as recent efforts for producing open datasets and benchmarks for the community.
fastMRI: An Open Dataset and Benchmarks for Accelerated MRI
Nov 21, 2018
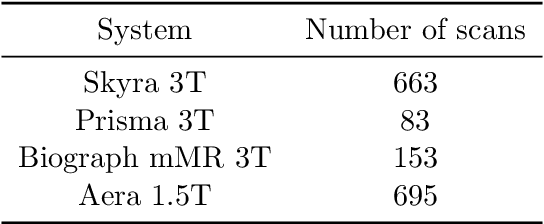
Abstract:Accelerating Magnetic Resonance Imaging (MRI) by taking fewer measurements has the potential to reduce medical costs, minimize stress to patients and make MRI possible in applications where it is currently prohibitively slow or expensive. We introduce the fastMRI dataset, a large-scale collection of both raw MR measurements and clinical MR images, that can be used for training and evaluation of machine-learning approaches to MR image reconstruction. By introducing standardized evaluation criteria and a freely-accessible dataset, our goal is to help the community make rapid advances in the state of the art for MR image reconstruction. We also provide a self-contained introduction to MRI for machine learning researchers with no medical imaging background.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge