Ravinder R. Regatte
Alternating Learning Approach for Variational Networks and Undersampling Pattern in Parallel MRI Applications
Oct 27, 2021
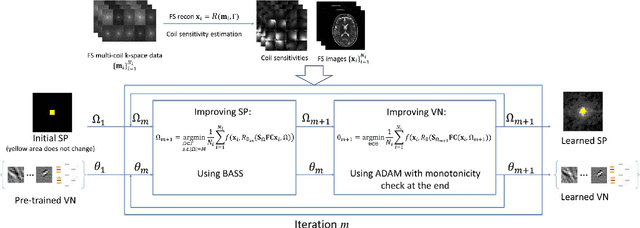


Abstract:Purpose: To propose an alternating learning approach to learn the sampling pattern (SP) and the parameters of variational networks (VN) in accelerated parallel magnetic resonance imaging (MRI). Methods: The approach alternates between improving the SP, using bias-accelerated subset selection, and improving parameters of the VN, using ADAM with monotonicity verification. The algorithm learns an effective pair: an SP that captures fewer k-space samples generating undersampling artifacts that are removed by the VN reconstruction. The proposed approach was tested for stability and convergence, considering different initial SPs. The quality of the VNs and SPs was compared against other approaches, including joint learning methods and VN learning with fixed variable density Poisson-disc SPs, using two different datasets and different acceleration factors (AF). Results: The root mean squared error (RMSE) improvements ranged from 14.9% to 51.2% considering AF from 2 to 20 in the tested brain and knee joint datasets when compared to the other approaches. The proposed approach has shown stable convergence, obtaining similar SPs with the same RMSE under different initial conditions. Conclusion: The proposed approach was stable and learned effective SPs with the corresponding VN parameters that produce images with better quality than other approaches, improving accelerated parallel MRI applications.
Fast Data-Driven Learning of MRI Sampling Pattern for Large Scale Problems
Nov 04, 2020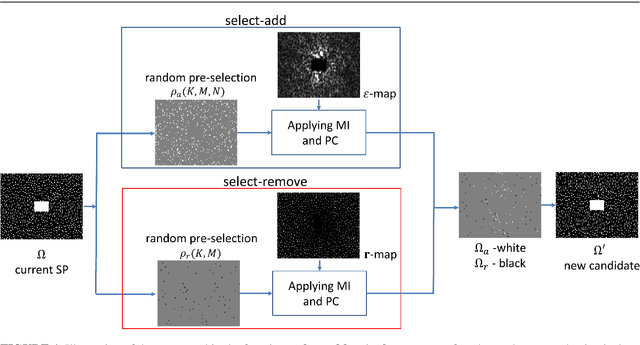
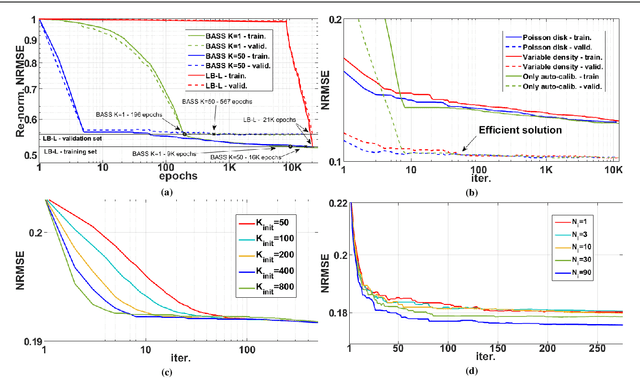

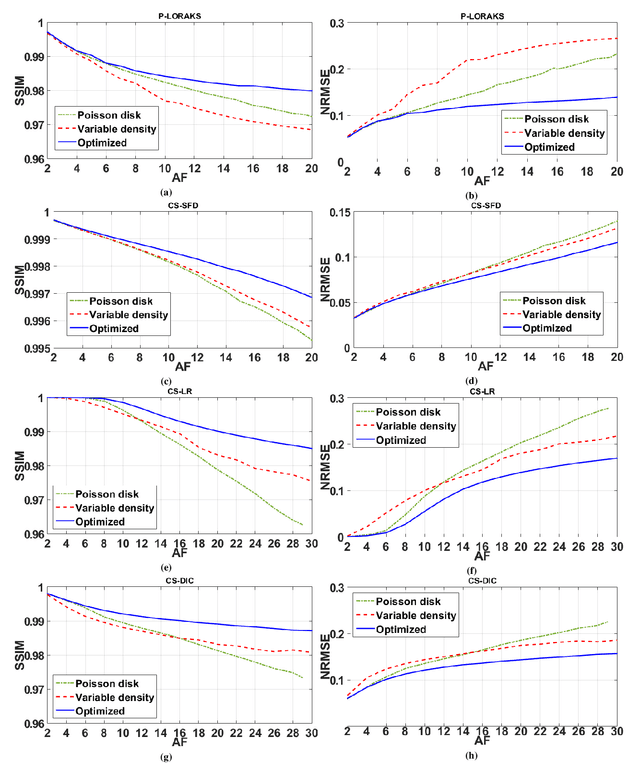
Abstract:Purpose: A fast data-driven optimization approach, named bias-accelerated subset selection (BASS), is proposed for learning efficacious sampling patterns (SPs) with the purpose of reducing scan time in large-dimensional parallel MRI. Methods: BASS is applicable when Cartesian fully-sampled k-space data of specific anatomy is available for training and the reconstruction method is specified, learning which k-space points are more relevant for the specific anatomy and reconstruction in recovering the non-sampled points. BASS was tested with four reconstruction methods for parallel MRI based on low-rankness and sparsity that allow a free choice of the SP. Two datasets were tested, one of the brain images for high-resolution imaging and another of knee images for quantitative mapping of the cartilage. Results: BASS, with its low computational cost and fast convergence, obtained SPs 100 times faster than the current best greedy approaches. Reconstruction quality increased up to 45\% with our learned SP over that provided by variable density and Poisson disk SPs, considering the same scan time. Optionally, the scan time can be nearly halved without loss of reconstruction quality. Conclusion: Compared with current approaches, BASS can be used to rapidly learn effective SPs for various reconstruction methods, using larger SP and larger datasets. This enables a better selection of efficacious sampling-reconstruction pairs for specific MRI problems.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge