Jeffrey Huang
Design and Performance Evaluation of an Elbow-Based Biomechanical Energy Harvester
Oct 11, 2024
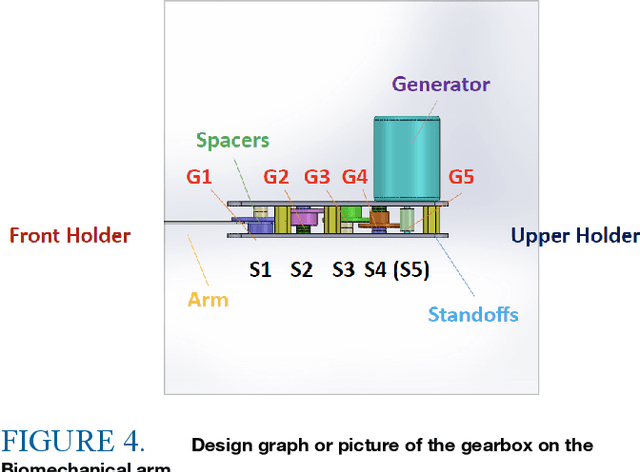
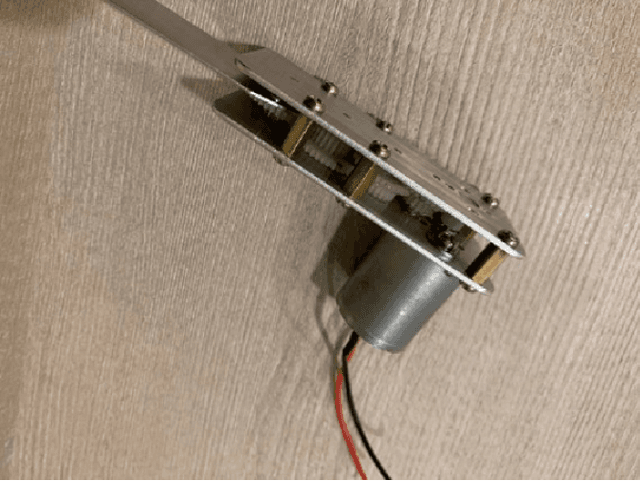
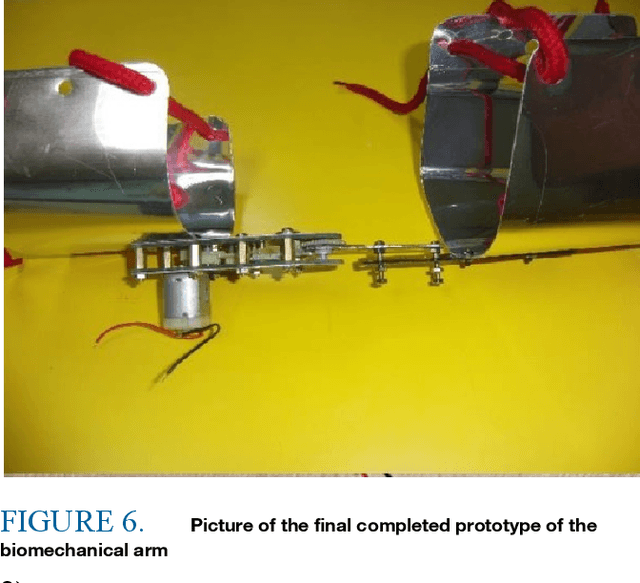
Abstract:Carbon emissions have long been attributed to the increase in climate change. With the effects of climate change escalating in the past few years, there has been an increased effort to find green alternatives to power generation, which has been a major contributor to carbon emissions. One prominent way that has arisen is biomechanical energy, or harvesting energy based on natural human movement. This study will evaluate the feasibility of electric generation using a gear and generator-based biomechanical energy harvester in the elbow joint. The joint was chosen using kinetic arm analysis through MediaPipe, in which the elbow joint showed much higher angular velocity during walking, thus showing more potential as a place to construct the harvester. Leg joints were excluded to not obstruct daily movement. The gear and generator type was decided to maximize energy production in the elbow joint. The device was constructed using a gearbox and a generator. The results show that it generated as much as 0.16 watts using the optimal resistance. This demonstrates the feasibility of electric generation with an elbow joint gear and generator-type biomechanical energy harvester.
Understanding Performance of Long-Document Ranking Models through Comprehensive Evaluation and Leaderboarding
Jul 04, 2022
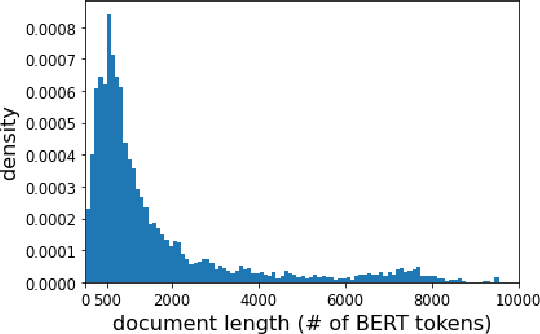
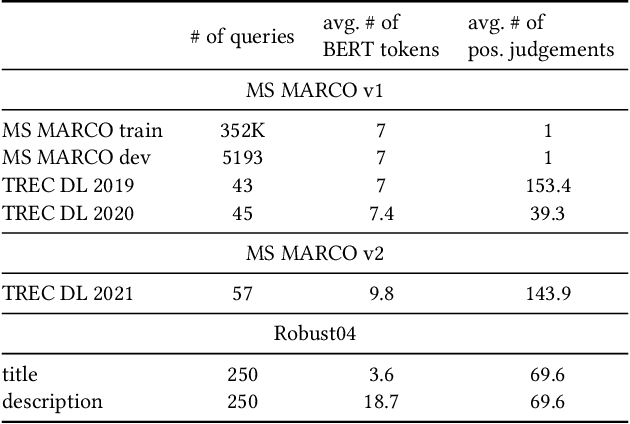
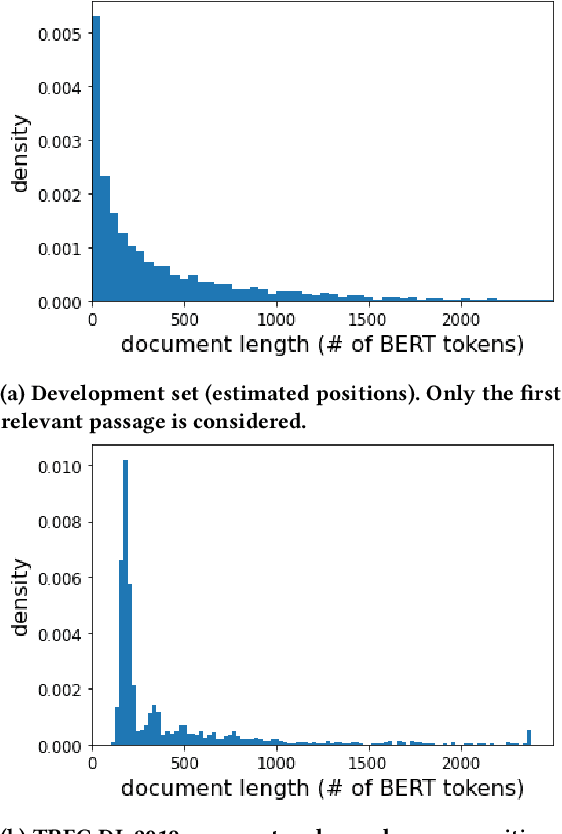
Abstract:We carry out a comprehensive evaluation of 13 recent models for ranking of long documents using two popular collections (MS MARCO documents and Robust04). Our model zoo includes two specialized Transformer models (such as Longformer) that can process long documents without the need to split them. Along the way, we document several difficulties regarding training and comparing such models. Somewhat surprisingly, we find the simple FirstP baseline (truncating documents to satisfy the input-sequence constraint of a typical Transformer model) to be quite effective. We analyze the distribution of relevant passages (inside documents) to explain this phenomenon. We further argue that, despite their widespread use, Robust04 and MS MARCO documents are not particularly useful for benchmarking of long-document models.
DARTS: DenseUnet-based Automatic Rapid Tool for brain Segmentation
Nov 14, 2019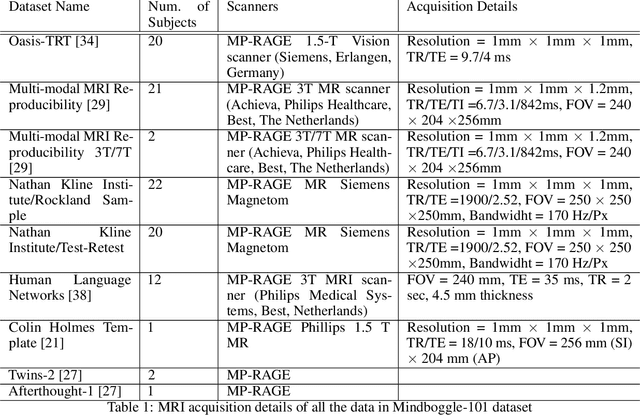
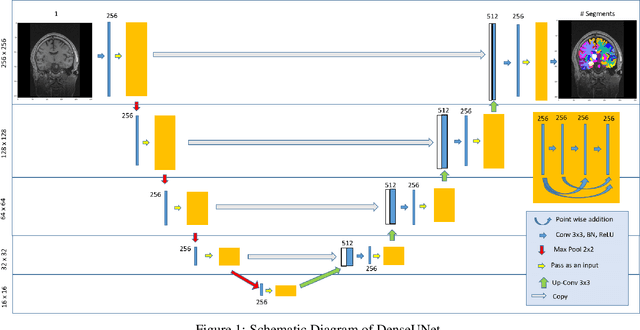
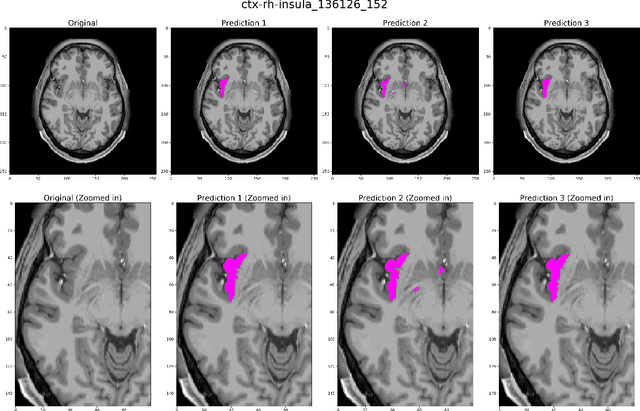
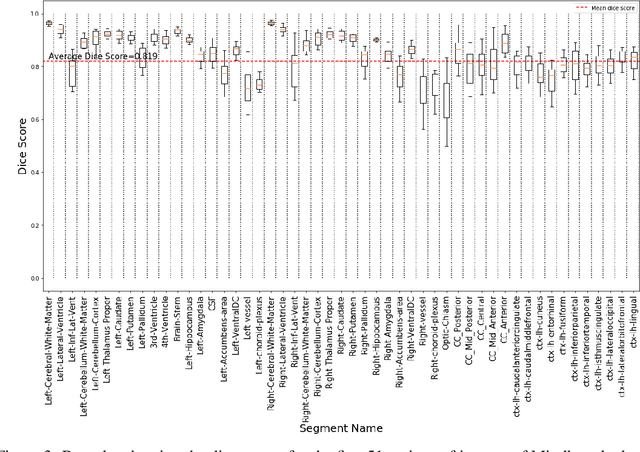
Abstract:Quantitative, volumetric analysis of Magnetic Resonance Imaging (MRI) is a fundamental way researchers study the brain in a host of neurological conditions including normal maturation and aging. Despite the availability of open-source brain segmentation software, widespread clinical adoption of volumetric analysis has been hindered due to processing times and reliance on manual corrections. Here, we extend the use of deep learning models from proof-of-concept, as previously reported, to present a comprehensive segmentation of cortical and deep gray matter brain structures matching the standard regions of aseg+aparc included in the commonly used open-source tool, Freesurfer. The work presented here provides a real-life, rapid deep learning-based brain segmentation tool to enable clinical translation as well as research application of quantitative brain segmentation. The advantages of the presented tool include short (~1 minute) processing time and improved segmentation quality. This is the first study to perform quick and accurate segmentation of 102 brain regions based on the surface-based protocol (DMK protocol), widely used by experts in the field. This is also the first work to include an expert reader study to assess the quality of the segmentation obtained using a deep-learning-based model. We show the superior performance of our deep-learning-based models over the traditional segmentation tool, Freesurfer. We refer to the proposed deep learning-based tool as DARTS (DenseUnet-based Automatic Rapid Tool for brain Segmentation). Our tool and trained models are available at https://github.com/NYUMedML/DARTS
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge