Hamza Javed
MEDIC: Towards a Comprehensive Framework for Evaluating LLMs in Clinical Applications
Sep 11, 2024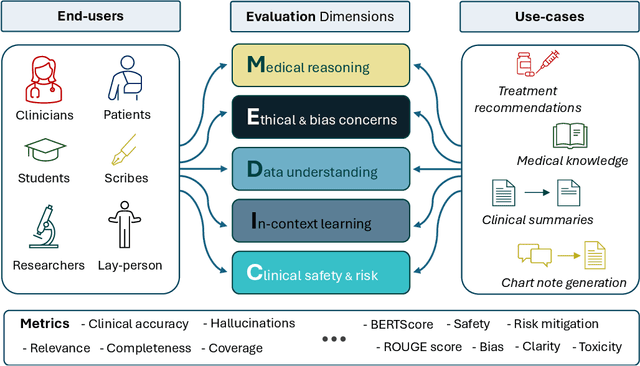
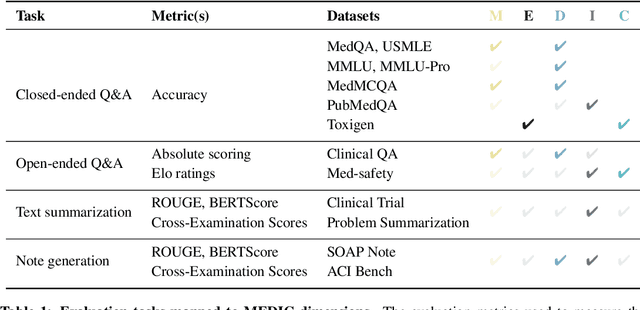
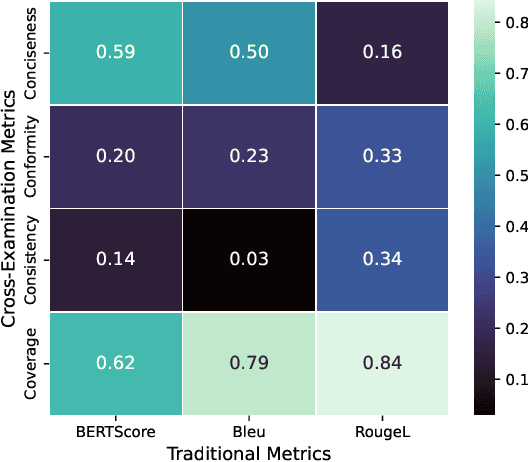
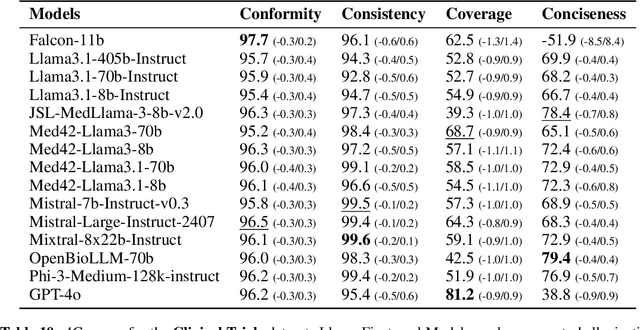
Abstract:The rapid development of Large Language Models (LLMs) for healthcare applications has spurred calls for holistic evaluation beyond frequently-cited benchmarks like USMLE, to better reflect real-world performance. While real-world assessments are valuable indicators of utility, they often lag behind the pace of LLM evolution, likely rendering findings obsolete upon deployment. This temporal disconnect necessitates a comprehensive upfront evaluation that can guide model selection for specific clinical applications. We introduce MEDIC, a framework assessing LLMs across five critical dimensions of clinical competence: medical reasoning, ethics and bias, data and language understanding, in-context learning, and clinical safety. MEDIC features a novel cross-examination framework quantifying LLM performance across areas like coverage and hallucination detection, without requiring reference outputs. We apply MEDIC to evaluate LLMs on medical question-answering, safety, summarization, note generation, and other tasks. Our results show performance disparities across model sizes, baseline vs medically finetuned models, and have implications on model selection for applications requiring specific model strengths, such as low hallucination or lower cost of inference. MEDIC's multifaceted evaluation reveals these performance trade-offs, bridging the gap between theoretical capabilities and practical implementation in healthcare settings, ensuring that the most promising models are identified and adapted for diverse healthcare applications.
Accelerating Recurrent Neural Networks for Gravitational Wave Experiments
Jun 26, 2021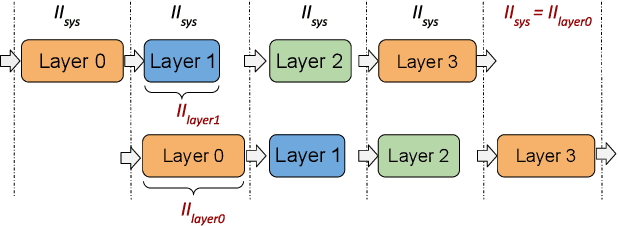
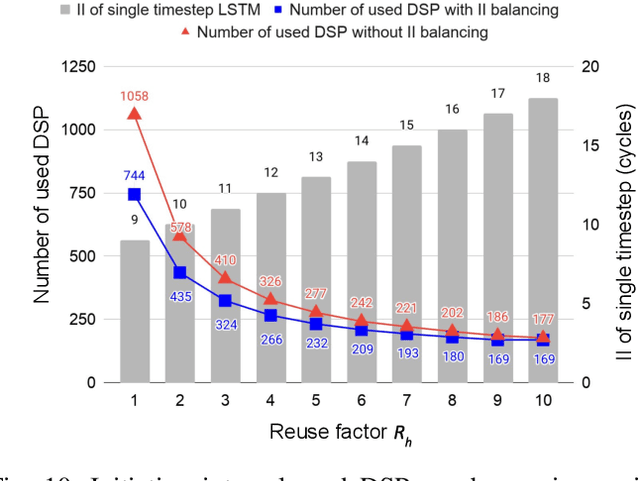
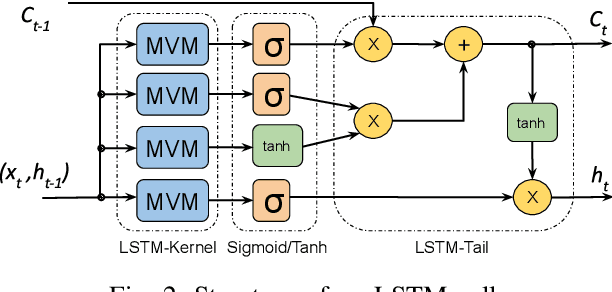
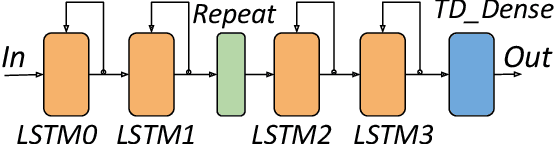
Abstract:This paper presents novel reconfigurable architectures for reducing the latency of recurrent neural networks (RNNs) that are used for detecting gravitational waves. Gravitational interferometers such as the LIGO detectors capture cosmic events such as black hole mergers which happen at unknown times and of varying durations, producing time-series data. We have developed a new architecture capable of accelerating RNN inference for analyzing time-series data from LIGO detectors. This architecture is based on optimizing the initiation intervals (II) in a multi-layer LSTM (Long Short-Term Memory) network, by identifying appropriate reuse factors for each layer. A customizable template for this architecture has been designed, which enables the generation of low-latency FPGA designs with efficient resource utilization using high-level synthesis tools. The proposed approach has been evaluated based on two LSTM models, targeting a ZYNQ 7045 FPGA and a U250 FPGA. Experimental results show that with balanced II, the number of DSPs can be reduced up to 42% while achieving the same IIs. When compared to other FPGA-based LSTM designs, our design can achieve about 4.92 to 12.4 times lower latency.
DeepMI: Deep Multi-lead ECG Fusion for Identifying Myocardial Infarction and its Occurrence-time
Mar 31, 2021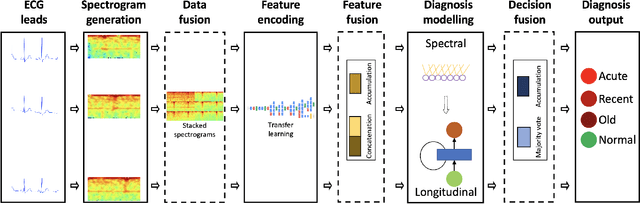
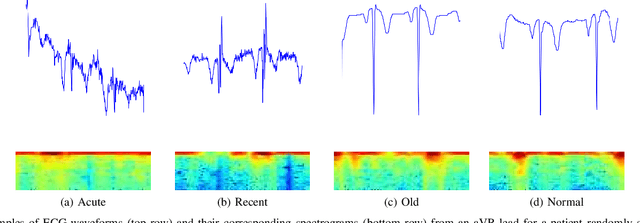
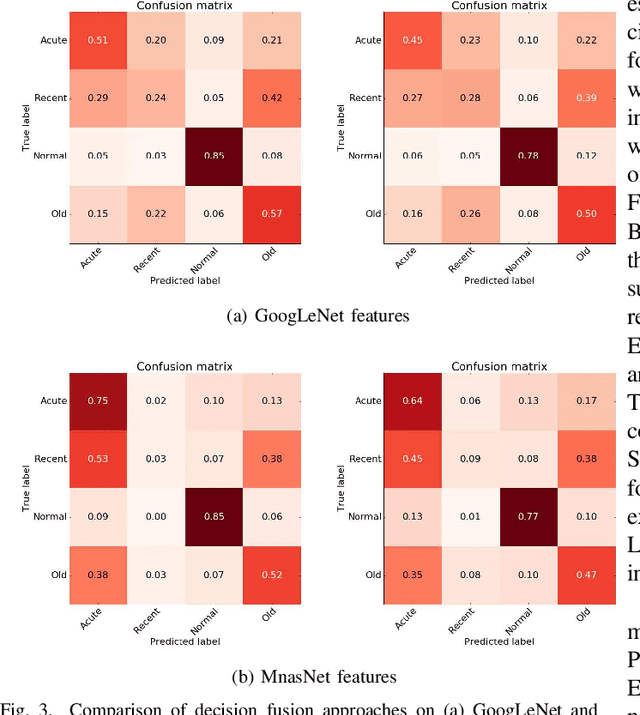
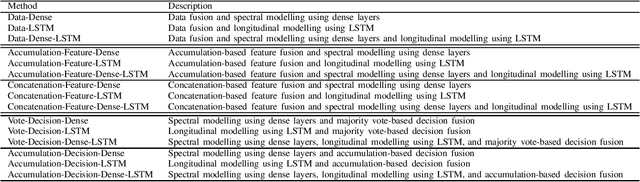
Abstract:Myocardial Infarction (MI) has the highest mortality of all cardiovascular diseases (CVDs). Detection of MI and information regarding its occurrence-time in particular, would enable timely interventions that may improve patient outcomes, thereby reducing the global rise in CVD deaths. Electrocardiogram (ECG) recordings are currently used to screen MI patients. However, manual inspection of ECGs is time-consuming and prone to subjective bias. Machine learning methods have been adopted for automated ECG diagnosis, but most approaches require extraction of ECG beats or consider leads independently of one another. We propose an end-to-end deep learning approach, DeepMI, to classify MI from normal cases as well as identifying the time-occurrence of MI (defined as acute, recent and old), using a collection of fusion strategies on 12 ECG leads at data-, feature-, and decision-level. In order to minimise computational overhead, we employ transfer learning using existing computer vision networks. Moreover, we use recurrent neural networks to encode the longitudinal information inherent in ECGs. We validated DeepMI on a dataset collected from 17,381 patients, in which over 323,000 samples were extracted per ECG lead. We were able to classify normal cases as well as acute, recent and old onset cases of MI, with AUROCs of 96.7%, 82.9%, 68.6% and 73.8%, respectively. We have demonstrated a multi-lead fusion approach to detect the presence and occurrence-time of MI. Our end-to-end framework provides flexibility for different levels of multi-lead ECG fusion and performs feature extraction via transfer learning.
hls4ml: An Open-Source Codesign Workflow to Empower Scientific Low-Power Machine Learning Devices
Mar 23, 2021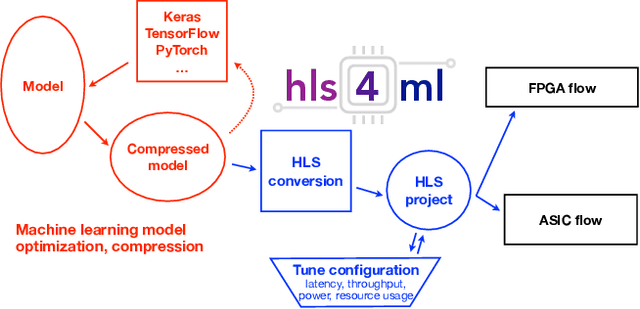
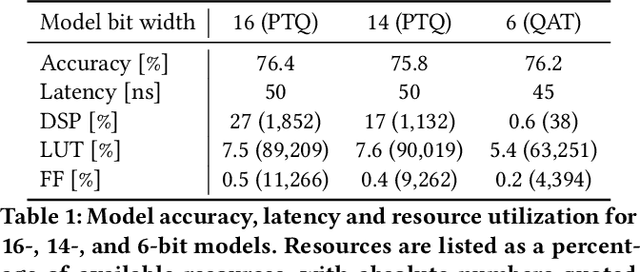
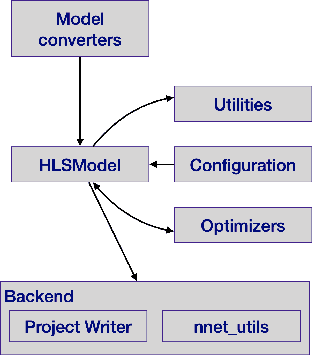
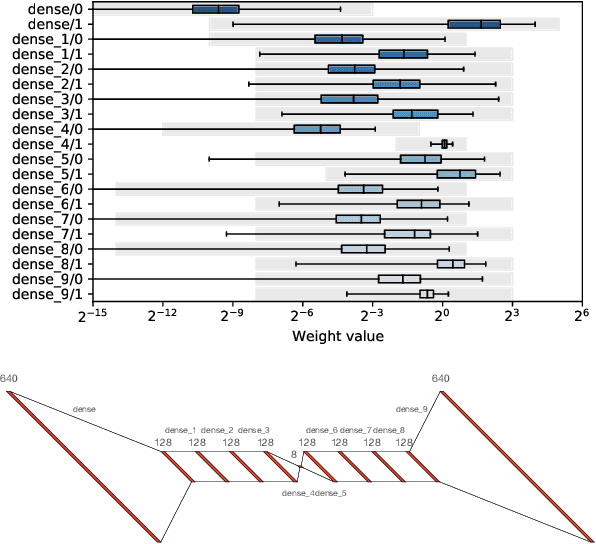
Abstract:Accessible machine learning algorithms, software, and diagnostic tools for energy-efficient devices and systems are extremely valuable across a broad range of application domains. In scientific domains, real-time near-sensor processing can drastically improve experimental design and accelerate scientific discoveries. To support domain scientists, we have developed hls4ml, an open-source software-hardware codesign workflow to interpret and translate machine learning algorithms for implementation with both FPGA and ASIC technologies. We expand on previous hls4ml work by extending capabilities and techniques towards low-power implementations and increased usability: new Python APIs, quantization-aware pruning, end-to-end FPGA workflows, long pipeline kernels for low power, and new device backends include an ASIC workflow. Taken together, these and continued efforts in hls4ml will arm a new generation of domain scientists with accessible, efficient, and powerful tools for machine-learning-accelerated discovery.
Severity Detection Tool for Patients with Infectious Disease
Dec 10, 2019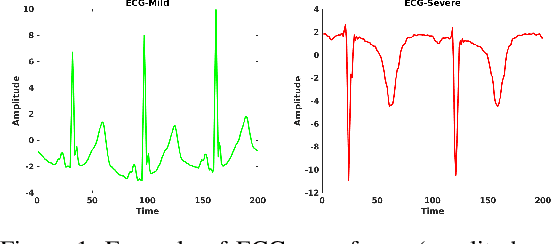
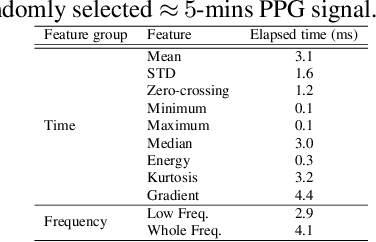
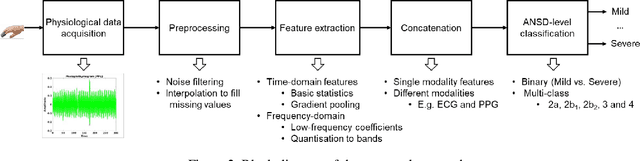
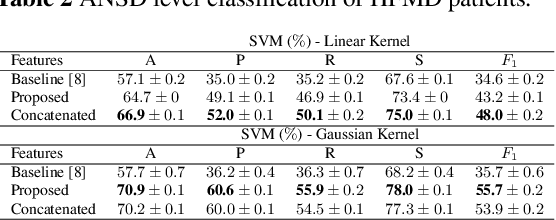
Abstract:Hand, foot and mouth disease (HFMD) and tetanus are serious infectious diseases in low and middle income countries. Tetanus in particular has a high mortality rate and its treatment is resource-demanding. Furthermore, HFMD often affects a large number of infants and young children. As a result, its treatment consumes enormous healthcare resources, especially when outbreaks occur. Autonomic nervous system dysfunction (ANSD) is the main cause of death for both HFMD and tetanus patients. However, early detection of ANSD is a difficult and challenging problem. In this paper, we aim to provide a proof-of-principle to detect the ANSD level automatically by applying machine learning techniques to physiological patient data, such as electrocardiogram (ECG) and photoplethysmogram (PPG) waveforms, which can be collected using low-cost wearable sensors. Efficient features are extracted that encode variations in the waveforms in the time and frequency domains. A support vector machine is employed to classify the ANSD levels. The proposed approach is validated on multiple datasets of HFMD and tetanus patients in Vietnam. Results show that encouraging performance is achieved in classifying ANSD levels. Moreover, the proposed features are simple, more generalisable and outperformed the standard heart rate variability (HRV) analysis. The proposed approach would facilitate both the diagnosis and treatment of infectious diseases in low and middle income countries, and thereby improve overall patient care.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge