Ahmed H. Shahin
A computationally frugal open-source foundation model for thoracic disease detection in lung cancer screening programs
Jul 02, 2025Abstract:Low-dose computed tomography (LDCT) imaging employed in lung cancer screening (LCS) programs is increasing in uptake worldwide. LCS programs herald a generational opportunity to simultaneously detect cancer and non-cancer-related early-stage lung disease. Yet these efforts are hampered by a shortage of radiologists to interpret scans at scale. Here, we present TANGERINE, a computationally frugal, open-source vision foundation model for volumetric LDCT analysis. Designed for broad accessibility and rapid adaptation, TANGERINE can be fine-tuned off the shelf for a wide range of disease-specific tasks with limited computational resources and training data. Relative to models trained from scratch, TANGERINE demonstrates fast convergence during fine-tuning, thereby requiring significantly fewer GPU hours, and displays strong label efficiency, achieving comparable or superior performance with a fraction of fine-tuning data. Pretrained using self-supervised learning on over 98,000 thoracic LDCTs, including the UK's largest LCS initiative to date and 27 public datasets, TANGERINE achieves state-of-the-art performance across 14 disease classification tasks, including lung cancer and multiple respiratory diseases, while generalising robustly across diverse clinical centres. By extending a masked autoencoder framework to 3D imaging, TANGERINE offers a scalable solution for LDCT analysis, departing from recent closed, resource-intensive models by combining architectural simplicity, public availability, and modest computational requirements. Its accessible, open-source lightweight design lays the foundation for rapid integration into next-generation medical imaging tools that could transform LCS initiatives, allowing them to pivot from a singular focus on lung cancer detection to comprehensive respiratory disease management in high-risk populations.
4D VQ-GAN: Synthesising Medical Scans at Any Time Point for Personalised Disease Progression Modelling of Idiopathic Pulmonary Fibrosis
Feb 08, 2025Abstract:Understanding the progression trajectories of diseases is crucial for early diagnosis and effective treatment planning. This is especially vital for life-threatening conditions such as Idiopathic Pulmonary Fibrosis (IPF), a chronic, progressive lung disease with a prognosis comparable to many cancers. Computed tomography (CT) imaging has been established as a reliable diagnostic tool for IPF. Accurately predicting future CT scans of early-stage IPF patients can aid in developing better treatment strategies, thereby improving survival outcomes. In this paper, we propose 4D Vector Quantised Generative Adversarial Networks (4D-VQ-GAN), a model capable of generating realistic CT volumes of IPF patients at any time point. The model is trained using a two-stage approach. In the first stage, a 3D-VQ-GAN is trained to reconstruct CT volumes. In the second stage, a Neural Ordinary Differential Equation (ODE) based temporal model is trained to capture the temporal dynamics of the quantised embeddings generated by the encoder in the first stage. We evaluate different configurations of our model for generating longitudinal CT scans and compare the results against ground truth data, both quantitatively and qualitatively. For validation, we conduct survival analysis using imaging biomarkers derived from generated CT scans and achieve a C-index comparable to that of biomarkers derived from the real CT scans. The survival analysis results demonstrate the potential clinical utility inherent to generated longitudinal CT scans, showing that they can reliably predict survival outcomes.
CenTime: Event-Conditional Modelling of Censoring in Survival Analysis
Sep 15, 2023Abstract:Survival analysis is a valuable tool for estimating the time until specific events, such as death or cancer recurrence, based on baseline observations. This is particularly useful in healthcare to prognostically predict clinically important events based on patient data. However, existing approaches often have limitations; some focus only on ranking patients by survivability, neglecting to estimate the actual event time, while others treat the problem as a classification task, ignoring the inherent time-ordered structure of the events. Furthermore, the effective utilization of censored samples - training data points where the exact event time is unknown - is essential for improving the predictive accuracy of the model. In this paper, we introduce CenTime, a novel approach to survival analysis that directly estimates the time to event. Our method features an innovative event-conditional censoring mechanism that performs robustly even when uncensored data is scarce. We demonstrate that our approach forms a consistent estimator for the event model parameters, even in the absence of uncensored data. Furthermore, CenTime is easily integrated with deep learning models with no restrictions on batch size or the number of uncensored samples. We compare our approach with standard survival analysis methods, including the Cox proportional-hazard model and DeepHit. Our results indicate that CenTime offers state-of-the-art performance in predicting time-to-death while maintaining comparable ranking performance. Our implementation is publicly available at https://github.com/ahmedhshahin/CenTime.
From Sparse to Precise: A Practical Editing Approach for Intracardiac Echocardiography Segmentation
Mar 20, 2023Abstract:Accurate and safe catheter ablation procedures for patients with atrial fibrillation require precise segmentation of cardiac structures in Intracardiac Echocardiography (ICE) imaging. Prior studies have suggested methods that employ 3D geometry information from the ICE transducer to create a sparse ICE volume by placing 2D frames in a 3D grid, enabling training of 3D segmentation models. However, the resulting 3D masks from these models can be inaccurate and may lead to serious clinical complications due to the sparse sampling in ICE data, frames misalignment, and cardiac motion. To address this issue, we propose an interactive editing framework that allows users to edit segmentation output by drawing scribbles on a 2D frame. The user interaction is mapped to the 3D grid and utilized to execute an editing step that modifies the segmentation in the vicinity of the interaction while preserving the previous segmentation away from the interaction. Furthermore, our framework accommodates multiple edits to the segmentation output in a sequential manner without compromising previous edits. This paper presents a novel loss function and a novel evaluation metric specifically designed for editing. Results from cross-validation and testing indicate that our proposed loss function outperforms standard losses and training strategies in terms of segmentation quality and following user input. Additionally, we show quantitatively and qualitatively that subsequent edits do not compromise previous edits when using our method, as opposed to standard segmentation losses. Overall, our approach enhances the accuracy of the segmentation while avoiding undesired changes away from user interactions and without compromising the quality of previously edited regions, leading to better patient outcomes.
Survival Analysis for Idiopathic Pulmonary Fibrosis using CT Images and Incomplete Clinical Data
Mar 21, 2022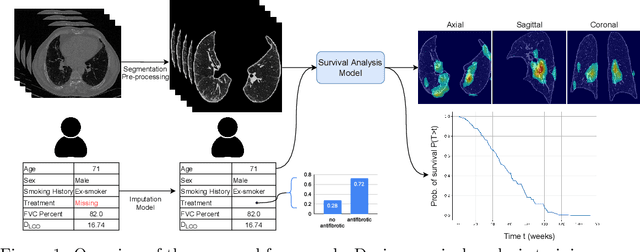
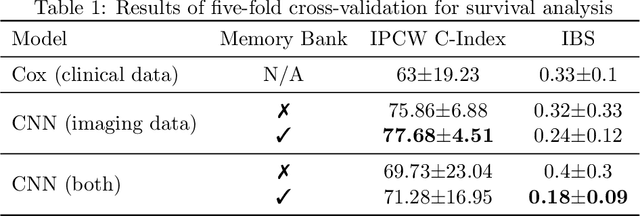
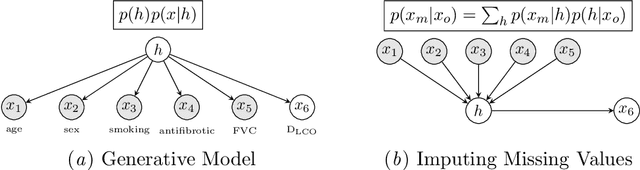

Abstract:Idiopathic Pulmonary Fibrosis (IPF) is an inexorably progressive fibrotic lung disease with a variable and unpredictable rate of progression. CT scans of the lungs inform clinical assessment of IPF patients and contain pertinent information related to disease progression. In this work, we propose a multi-modal method that uses neural networks and memory banks to predict the survival of IPF patients using clinical and imaging data. The majority of clinical IPF patient records have missing data (e.g. missing lung function tests). To this end, we propose a probabilistic model that captures the dependencies between the observed clinical variables and imputes missing ones. This principled approach to missing data imputation can be naturally combined with a deep survival analysis model. We show that the proposed framework yields significantly better survival analysis results than baselines in terms of concordance index and integrated Brier score. Our work also provides insights into novel image-based biomarkers that are linked to mortality.
FAIRS -- Soft Focus Generator and Attention for Robust Object Segmentation from Extreme Points
Apr 04, 2020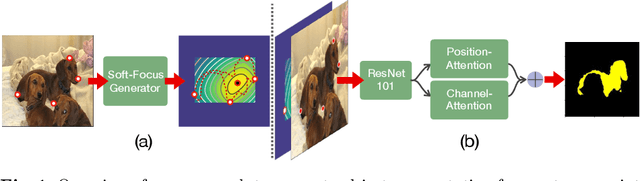
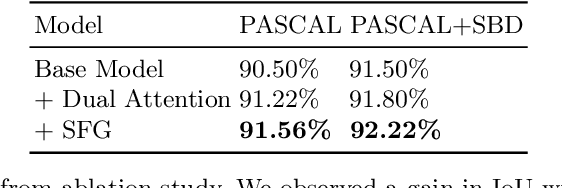
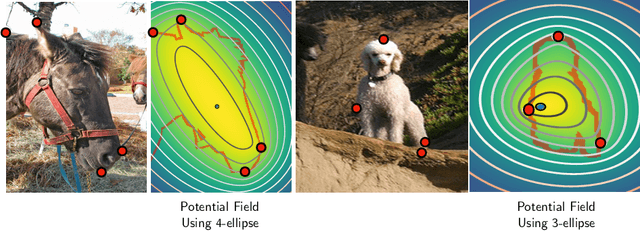
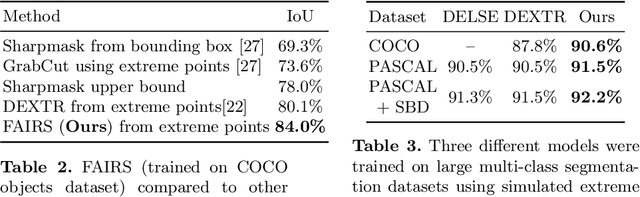
Abstract:Semantic segmentation from user inputs has been actively studied to facilitate interactive segmentation for data annotation and other applications. Recent studies have shown that extreme points can be effectively used to encode user inputs. A heat map generated from the extreme points can be appended to the RGB image and input to the model for training. In this study, we present FAIRS -- a new approach to generate object segmentation from user inputs in the form of extreme points and corrective clicks. We propose a novel approach for effectively encoding the user input from extreme points and corrective clicks, in a novel and scalable manner that allows the network to work with a variable number of clicks, including corrective clicks for output refinement. We also integrate a dual attention module with our approach to increase the efficacy of the model in preferentially attending to the objects. We demonstrate that these additions help achieve significant improvements over state-of-the-art in dense object segmentation from user inputs, on multiple large-scale datasets. Through experiments, we demonstrate our method's ability to generate high-quality training data as well as its scalability in incorporating extreme points, guiding clicks, and corrective clicks in a principled manner.
Extreme Points Derived Confidence Map as a Cue For Class-Agnostic Segmentation Using Deep Neural Network
Jun 06, 2019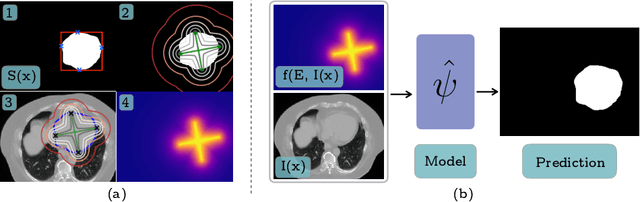
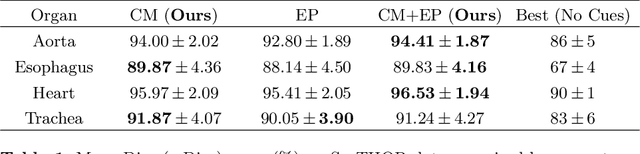
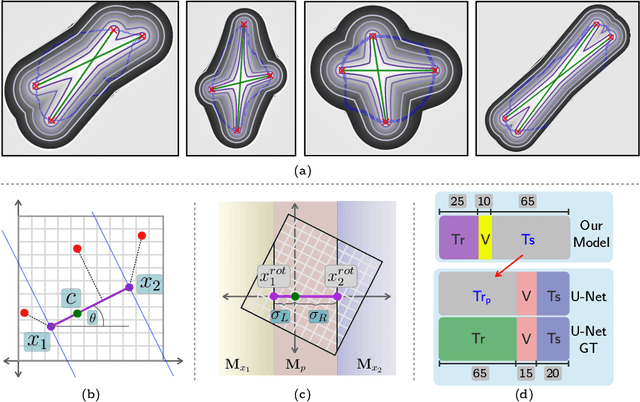
Abstract:To automate the process of segmenting an anatomy of interest, we can learn a model from previously annotated data. The learning-based approach uses annotations to train a model that tries to emulate the expert labeling on a new data set. While tremendous progress has been made using such approaches, labeling of medical images remains a time-consuming and expensive task. In this paper, we evaluate the utility of extreme points in learning to segment. Specifically, we propose a novel approach to compute a confidence map from extreme points that quantitatively encodes the priors derived from extreme points. We use the confidence map as a cue to train a deep neural network based on ResNet-101 and PSP module to develop a class-agnostic segmentation model that outperforms state-of-the-art method that employs extreme points as a cue. Further, we evaluate a realistic use-case by using our model to generate training data for supervised learning (U-Net) and observed that U-Net performs comparably when trained with either the generated data or the ground truth data. These findings suggest that models trained using cues can be used to generate reliable training data.
Deep Convolutional Encoder-Decoders with Aggregated Multi-Resolution Skip Connections for Skin Lesion Segmentation
Jan 26, 2019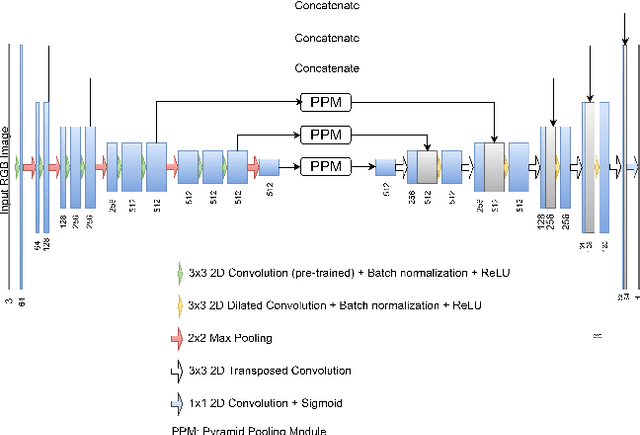
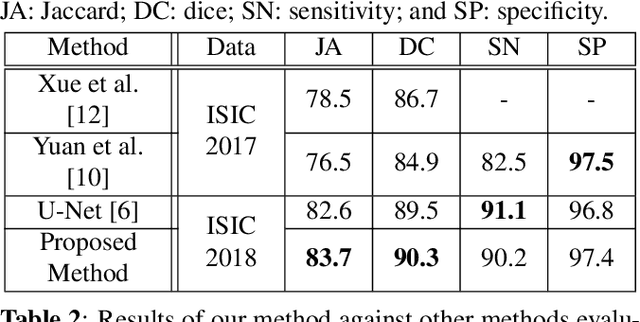
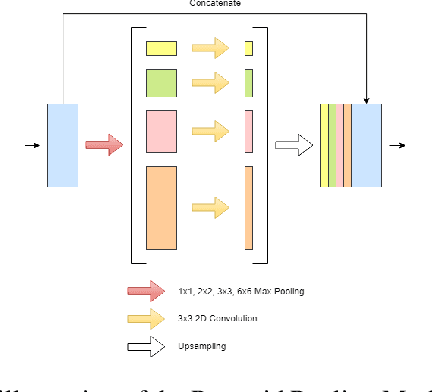
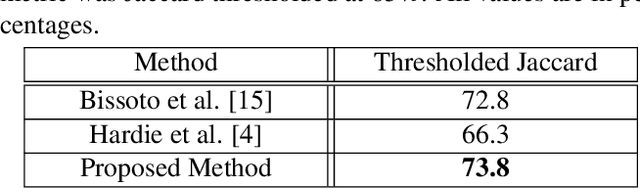
Abstract:The prevalence of skin melanoma is rapidly increasing as well as the recorded death cases of its patients. Automatic image segmentation tools play an important role in providing standardized computer-assisted analysis for skin melanoma patients. Current state-of-the-art segmentation methods are based on fully convolutional neural networks, which utilize an encoder-decoder approach. However, these methods produce coarse segmentation masks due to the loss of location information during the encoding layers. Inspired by Pyramid Scene Parsing Network (PSP-Net), we propose an encoder-decoder model that utilizes pyramid pooling modules in the deep skip connections which aggregate the global context and compensate for the lost spatial information. We trained and validated our approach using ISIC 2018: Skin Lesion Analysis Towards Melanoma Detection grand challenge dataset. Our approach showed a validation accuracy with a Jaccard index of 0.837, which outperforms U-Net. We believe that with this reported reliable accuracy, this method can be introduced for clinical practice.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge