Moucheng Xu
4D VQ-GAN: Synthesising Medical Scans at Any Time Point for Personalised Disease Progression Modelling of Idiopathic Pulmonary Fibrosis
Feb 08, 2025Abstract:Understanding the progression trajectories of diseases is crucial for early diagnosis and effective treatment planning. This is especially vital for life-threatening conditions such as Idiopathic Pulmonary Fibrosis (IPF), a chronic, progressive lung disease with a prognosis comparable to many cancers. Computed tomography (CT) imaging has been established as a reliable diagnostic tool for IPF. Accurately predicting future CT scans of early-stage IPF patients can aid in developing better treatment strategies, thereby improving survival outcomes. In this paper, we propose 4D Vector Quantised Generative Adversarial Networks (4D-VQ-GAN), a model capable of generating realistic CT volumes of IPF patients at any time point. The model is trained using a two-stage approach. In the first stage, a 3D-VQ-GAN is trained to reconstruct CT volumes. In the second stage, a Neural Ordinary Differential Equation (ODE) based temporal model is trained to capture the temporal dynamics of the quantised embeddings generated by the encoder in the first stage. We evaluate different configurations of our model for generating longitudinal CT scans and compare the results against ground truth data, both quantitatively and qualitatively. For validation, we conduct survival analysis using imaging biomarkers derived from generated CT scans and achieve a C-index comparable to that of biomarkers derived from the real CT scans. The survival analysis results demonstrate the potential clinical utility inherent to generated longitudinal CT scans, showing that they can reliably predict survival outcomes.
MRI Parameter Mapping via Gaussian Mixture VAE: Breaking the Assumption of Independent Pixels
Nov 16, 2024



Abstract:We introduce and demonstrate a new paradigm for quantitative parameter mapping in MRI. Parameter mapping techniques, such as diffusion MRI and quantitative MRI, have the potential to robustly and repeatably measure biologically-relevant tissue maps that strongly relate to underlying microstructure. Quantitative maps are calculated by fitting a model to multiple images, e.g. with least-squares or machine learning. However, the overwhelming majority of model fitting techniques assume that each voxel is independent, ignoring any co-dependencies in the data. This makes model fitting sensitive to voxelwise measurement noise, hampering reliability and repeatability. We propose a self-supervised deep variational approach that breaks the assumption of independent pixels, leveraging redundancies in the data to effectively perform data-driven regularisation of quantitative maps. We demonstrate that our approach outperforms current model fitting techniques in dMRI simulations and real data. Especially with a Gaussian mixture prior, our model enables sharper quantitative maps, revealing finer anatomical details that are not presented in the baselines. Our approach can hence support the clinical adoption of parameter mapping methods such as dMRI and qMRI.
In-Context Ensemble Improves Video-Language Models for Low-Level Workflow Understanding from Human Demonstrations
Sep 26, 2024



Abstract:A Standard Operating Procedure (SOP) defines a low-level, step-by-step written guide for a business software workflow based on a video demonstration. SOPs are a crucial step toward automating end-to-end software workflows. Manually creating SOPs can be time-consuming. Recent advancements in large video-language models offer the potential for automating SOP generation by analyzing recordings of human demonstrations. However, current large video-language models face challenges with zero-shot SOP generation. We explore in-context learning with video-language models for SOP generation. We report that in-context learning sometimes helps video-language models at SOP generation. We then propose an in-context ensemble learning to further enhance the capabilities of the models in SOP generation.
VAFO-Loss: VAscular Feature Optimised Loss Function for Retinal Artery/Vein Segmentation
Mar 12, 2022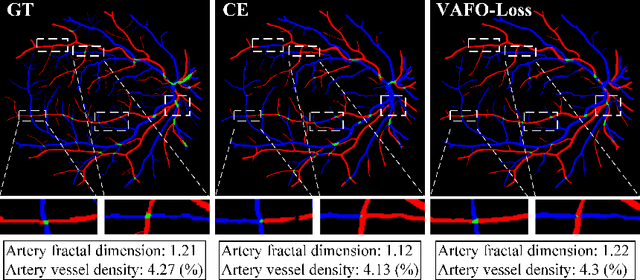
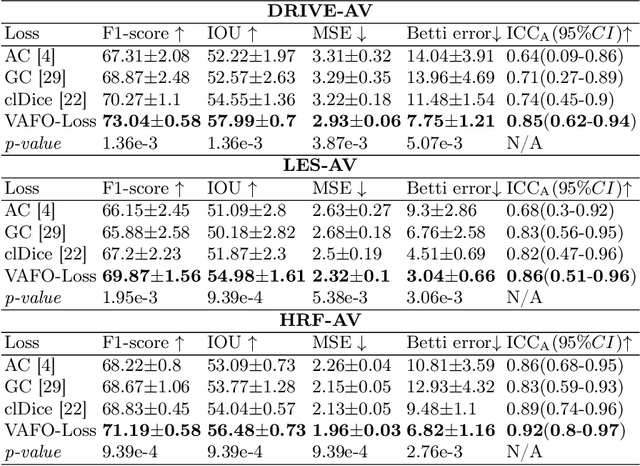
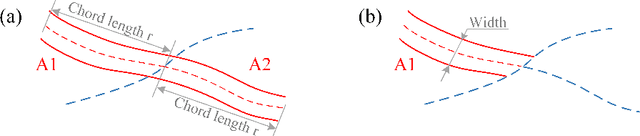
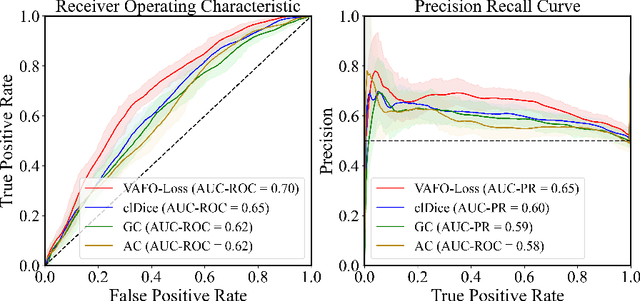
Abstract:Estimating clinically-relevant vascular features following vessel segmentation is a standard pipeline for retinal vessel analysis, which provides potential ocular biomarkers for both ophthalmic disease and systemic disease. In this work, we integrate these clinical features into a novel vascular feature optimised loss function (VAFO-Loss), in order to regularise networks to produce segmentation maps, with which more accurate vascular features can be derived. Two common vascular features, vessel density and fractal dimension, are identified to be sensitive to intra-segment misclassification, which is a well-recognised problem in multi-class artery/vein segmentation particularly hindering the estimation of these vascular features. Thus we encode these two features into VAFO-Loss. We first show that incorporating our end-to-end VAFO-Loss in standard segmentation networks indeed improves vascular feature estimation, yielding quantitative improvement in stroke incidence prediction, a clinical downstream task. We also report a technically interesting finding that the trained segmentation network, albeit biased by the feature optimised loss VAFO-Loss, shows statistically significant improvement in segmentation metrics, compared to those trained with other state-of-the-art segmentation losses.
Learning to Address Intra-segment Misclassification in Retinal Imaging
Apr 25, 2021



Abstract:Accurate multi-class segmentation is a long-standing challenge in medical imaging, especially in scenarios where classes share strong similarity. Segmenting retinal blood vessels in retinal photographs is one such scenario, in which arteries and veins need to be identified and differentiated from each other and from the background. Intra-segment misclassification, i.e. veins classified as arteries or vice versa, frequently occurs when arteries and veins intersect, whereas in binary retinal vessel segmentation, error rates are much lower. We thus propose a new approach that decomposes multi-class segmentation into multiple binary, followed by a binary-to-multi-class fusion network. The network merges representations of artery, vein, and multi-class feature maps, each of which are supervised by expert vessel annotation in adversarial training. A skip-connection based merging process explicitly maintains class-specific gradients to avoid gradient vanishing in deep layers, to favor the discriminative features. The results show that, our model respectively improves F1-score by 4.4\%, 5.1\%, and 4.2\% compared with three state-of-the-art deep learning based methods on DRIVE-AV, LES-AV, and HRF-AV data sets.
Foveation for Segmentation of Ultra-High Resolution Images
Jul 31, 2020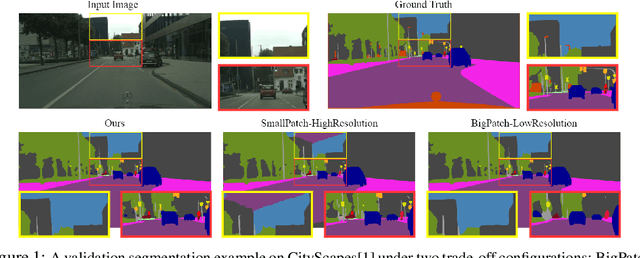

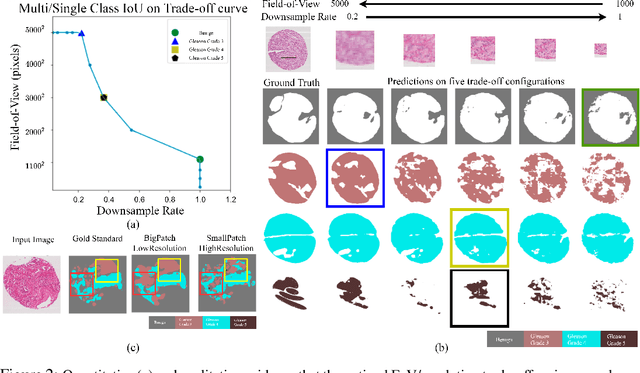
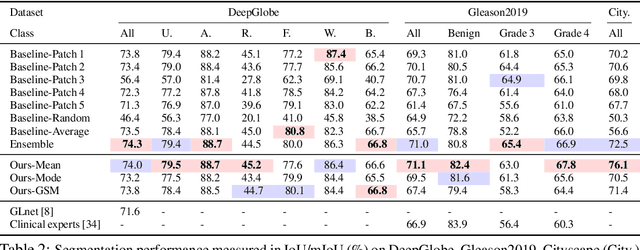
Abstract:Segmentation of ultra-high resolution images is challenging because of their enormous size, consisting of millions or even billions of pixels. Typical solutions include dividing input images into patches of fixed size and/or down-sampling to meet memory constraints. Such operations incur information loss in the field-of-view (FoV) i.e., spatial coverage and the image resolution. The impact on segmentation performance is, however, as yet understudied. In this work, we start with a motivational experiment which demonstrates that the trade-off between FoV and resolution affects the segmentation performance on ultra-high resolution images---and furthermore, its influence also varies spatially according to the local patterns in different areas. We then introduce foveation module, a learnable "dataloader" which, for a given ultra-high resolution image, adaptively chooses the appropriate configuration (FoV/resolution trade-off) of the input patch to feed to the downstream segmentation model at each spatial location of the image. The foveation module is jointly trained with the segmentation network to maximise the task performance. We demonstrate on three publicly available high-resolution image datasets that the foveation module consistently improves segmentation performance over the cases trained with patches of fixed FoV/resolution trade-off. Our approach achieves the SoTA performance on the DeepGlobe aerial image dataset. On the Gleason2019 histopathology dataset, our model achieves better segmentation accuracy for the two most clinically important and ambiguous classes (Gleason Grade 3 and 4) than the top performers in the challenge by 13.1% and 7.5%, and improves on the average performance of 6 human experts by 6.5% and 7.5%. Our code and trained models are available at $\text{https://github.com/lxasqjc/Foveation-Segmentation}$.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge