Tobias Goodwin-Allcock
MRI Parameter Mapping via Gaussian Mixture VAE: Breaking the Assumption of Independent Pixels
Nov 16, 2024



Abstract:We introduce and demonstrate a new paradigm for quantitative parameter mapping in MRI. Parameter mapping techniques, such as diffusion MRI and quantitative MRI, have the potential to robustly and repeatably measure biologically-relevant tissue maps that strongly relate to underlying microstructure. Quantitative maps are calculated by fitting a model to multiple images, e.g. with least-squares or machine learning. However, the overwhelming majority of model fitting techniques assume that each voxel is independent, ignoring any co-dependencies in the data. This makes model fitting sensitive to voxelwise measurement noise, hampering reliability and repeatability. We propose a self-supervised deep variational approach that breaks the assumption of independent pixels, leveraging redundancies in the data to effectively perform data-driven regularisation of quantitative maps. We demonstrate that our approach outperforms current model fitting techniques in dMRI simulations and real data. Especially with a Gaussian mixture prior, our model enables sharper quantitative maps, revealing finer anatomical details that are not presented in the baselines. Our approach can hence support the clinical adoption of parameter mapping methods such as dMRI and qMRI.
Patch-CNN: Training data-efficient deep learning for high-fidelity diffusion tensor estimation from minimal diffusion protocols
Jul 03, 2023Abstract:We propose a new method, Patch-CNN, for diffusion tensor (DT) estimation from only six-direction diffusion weighted images (DWI). Deep learning-based methods have been recently proposed for dMRI parameter estimation, using either voxel-wise fully-connected neural networks (FCN) or image-wise convolutional neural networks (CNN). In the acute clinical context -- where pressure of time limits the number of imaged directions to a minimum -- existing approaches either require an infeasible number of training images volumes (image-wise CNNs), or do not estimate the fibre orientations (voxel-wise FCNs) required for tractogram estimation. To overcome these limitations, we propose Patch-CNN, a neural network with a minimal (non-voxel-wise) convolutional kernel (3$\times$3$\times$3). Compared with voxel-wise FCNs, this has the advantage of allowing the network to leverage local anatomical information. Compared with image-wise CNNs, the minimal kernel vastly reduces training data demand. Evaluated against both conventional model fitting and a voxel-wise FCN, Patch-CNN, trained with a single subject is shown to improve the estimation of both scalar dMRI parameters and fibre orientation from six-direction DWIs. The improved fibre orientation estimation is shown to produce improved tractogram.
How can spherical CNNs benefit ML-based diffusion MRI parameter estimation?
Jul 01, 2022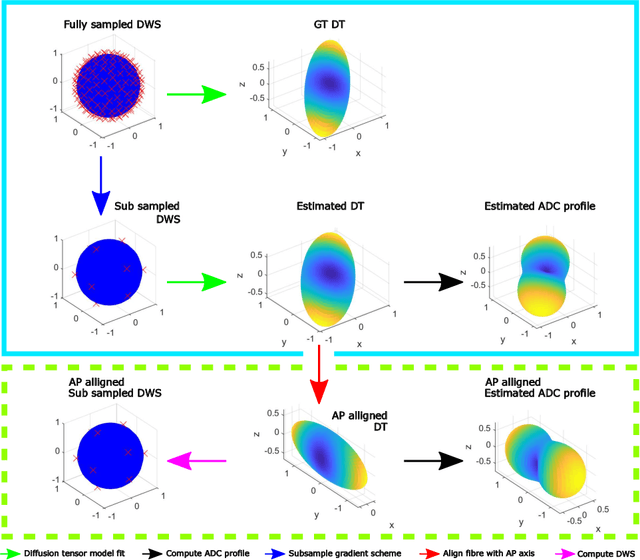
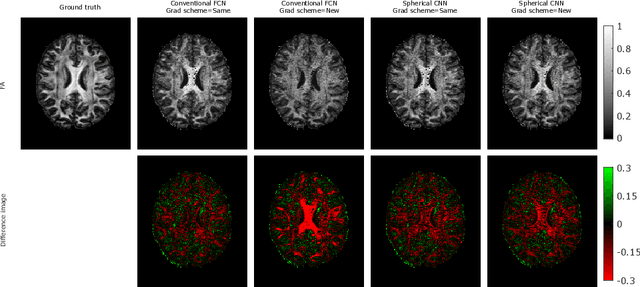
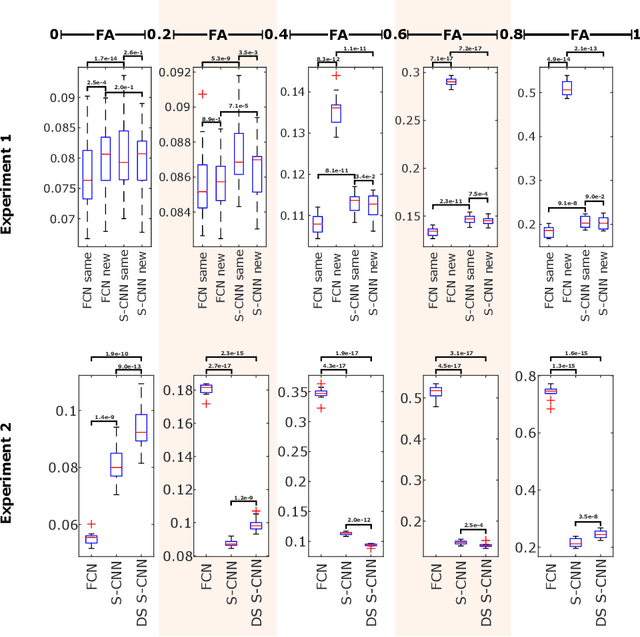
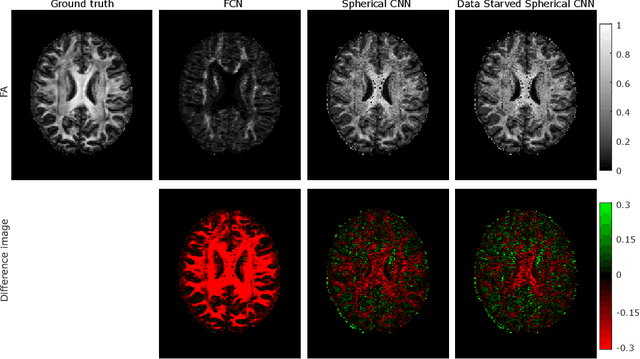
Abstract:This paper demonstrates spherical convolutional neural networks (S-CNN) offer distinct advantages over conventional fully-connected networks (FCN) at estimating scalar parameters of tissue microstructure from diffusion MRI (dMRI). Such microstructure parameters are valuable for identifying pathology and quantifying its extent. However, current clinical practice commonly acquires dMRI data consisting of only 6 diffusion weighted images (DWIs), limiting the accuracy and precision of estimated microstructure indices. Machine learning (ML) has been proposed to address this challenge. However, existing ML-based methods are not robust to differing dMRI gradient sampling schemes, nor are they rotation equivariant. Lack of robustness to sampling schemes requires a new network to be trained for each scheme, complicating the analysis of data from multiple sources. A possible consequence of the lack of rotational equivariance is that the training dataset must contain a diverse range of microstucture orientations. Here, we show spherical CNNs represent a compelling alternative that is robust to new sampling schemes as well as offering rotational equivariance. We show the latter can be leveraged to decrease the number of training datapoints required.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge