Anees Ur Rehman Hashmi
AnatomiX, an Anatomy-Aware Grounded Multimodal Large Language Model for Chest X-Ray Interpretation
Jan 06, 2026Abstract:Multimodal medical large language models have shown impressive progress in chest X-ray interpretation but continue to face challenges in spatial reasoning and anatomical understanding. Although existing grounding techniques improve overall performance, they often fail to establish a true anatomical correspondence, resulting in incorrect anatomical understanding in the medical domain. To address this gap, we introduce AnatomiX, a multitask multimodal large language model explicitly designed for anatomically grounded chest X-ray interpretation. Inspired by the radiological workflow, AnatomiX adopts a two stage approach: first, it identifies anatomical structures and extracts their features, and then leverages a large language model to perform diverse downstream tasks such as phrase grounding, report generation, visual question answering, and image understanding. Extensive experiments across multiple benchmarks demonstrate that AnatomiX achieves superior anatomical reasoning and delivers over 25% improvement in performance on anatomy grounding, phrase grounding, grounded diagnosis and grounded captioning tasks compared to existing approaches. Code and pretrained model are available at https://github.com/aneesurhashmi/anatomix
Continual Learning in Medical Imaging from Theory to Practice: A Survey and Practical Analysis
May 22, 2024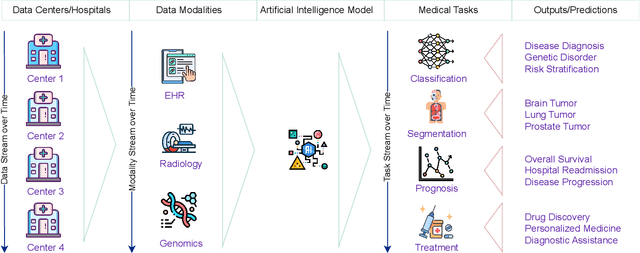
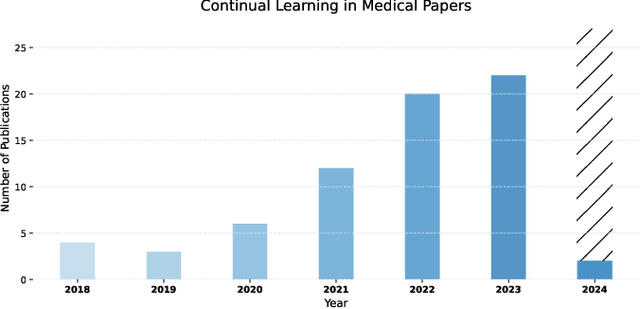

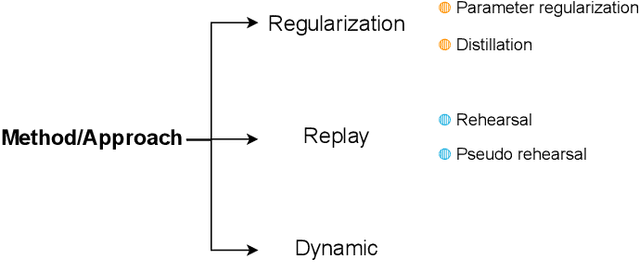
Abstract:Deep Learning has shown great success in reshaping medical imaging, yet it faces numerous challenges hindering widespread application. Issues like catastrophic forgetting and distribution shifts in the continuously evolving data stream increase the gap between research and applications. Continual Learning offers promise in addressing these hurdles by enabling the sequential acquisition of new knowledge without forgetting previous learnings in neural networks. In this survey, we comprehensively review the recent literature on continual learning in the medical domain, highlight recent trends, and point out the practical issues. Specifically, we survey the continual learning studies on classification, segmentation, detection, and other tasks in the medical domain. Furthermore, we develop a taxonomy for the reviewed studies, identify the challenges, and provide insights to overcome them. We also critically discuss the current state of continual learning in medical imaging, including identifying open problems and outlining promising future directions. We hope this survey will provide researchers with a useful overview of the developments in the field and will further increase interest in the community. To keep up with the fast-paced advancements in this field, we plan to routinely update the repository with the latest relevant papers at https://github.com/BioMedIA-MBZUAI/awesome-cl-in-medical .
On Enhancing Brain Tumor Segmentation Across Diverse Populations with Convolutional Neural Networks
May 05, 2024

Abstract:Brain tumor segmentation is a fundamental step in assessing a patient's cancer progression. However, manual segmentation demands significant expert time to identify tumors in 3D multimodal brain MRI scans accurately. This reliance on manual segmentation makes the process prone to intra- and inter-observer variability. This work proposes a brain tumor segmentation method as part of the BraTS-GoAT challenge. The task is to segment tumors in brain MRI scans automatically from various populations, such as adults, pediatrics, and underserved sub-Saharan Africa. We employ a recent CNN architecture for medical image segmentation, namely MedNeXt, as our baseline, and we implement extensive model ensembling and postprocessing for inference. Our experiments show that our method performs well on the unseen validation set with an average DSC of 85.54% and HD95 of 27.88. The code is available on https://github.com/BioMedIA-MBZUAI/BraTS2024_BioMedIAMBZ.
DynaMMo: Dynamic Model Merging for Efficient Class Incremental Learning for Medical Images
Apr 22, 2024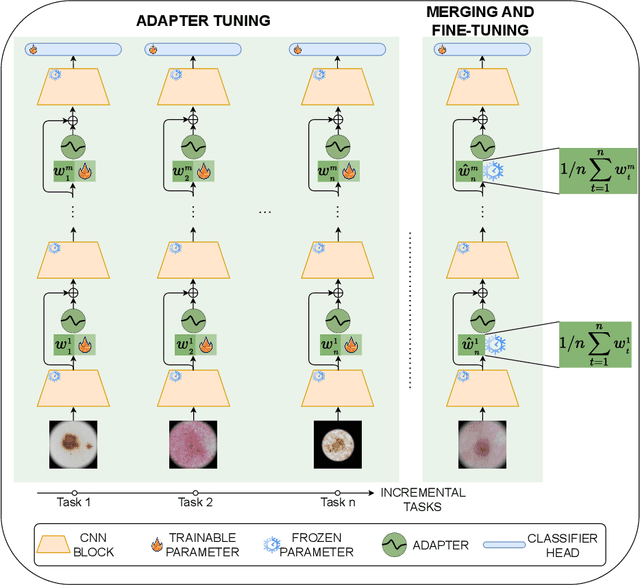
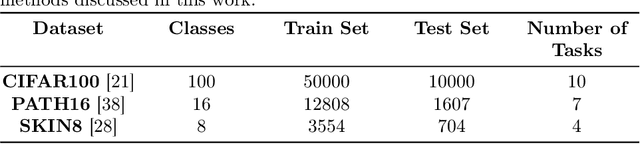
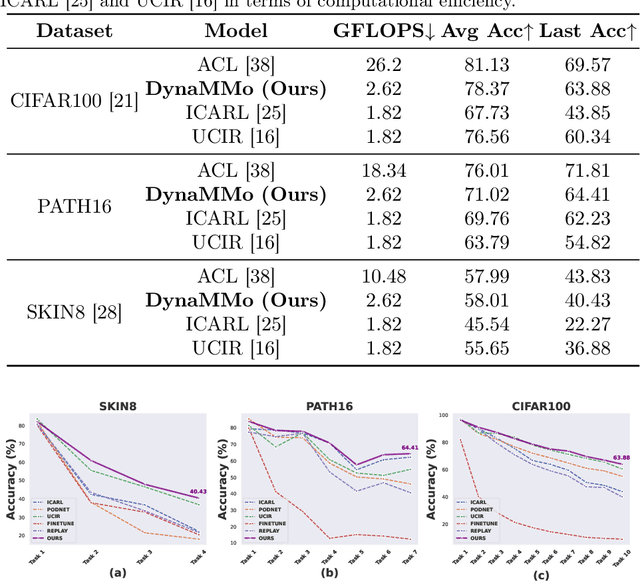
Abstract:Continual learning, the ability to acquire knowledge from new data while retaining previously learned information, is a fundamental challenge in machine learning. Various approaches, including memory replay, knowledge distillation, model regularization, and dynamic network expansion, have been proposed to address this issue. Thus far, dynamic network expansion methods have achieved state-of-the-art performance at the cost of incurring significant computational overhead. This is due to the need for additional model buffers, which makes it less feasible in resource-constrained settings, particularly in the medical domain. To overcome this challenge, we propose Dynamic Model Merging, DynaMMo, a method that merges multiple networks at different stages of model training to achieve better computational efficiency. Specifically, we employ lightweight learnable modules for each task and combine them into a unified model to minimize computational overhead. DynaMMo achieves this without compromising performance, offering a cost-effective solution for continual learning in medical applications. We evaluate DynaMMo on three publicly available datasets, demonstrating its effectiveness compared to existing approaches. DynaMMo offers around 10-fold reduction in GFLOPS with a small drop of 2.76 in average accuracy when compared to state-of-the-art dynamic-based approaches. The code implementation of this work will be available upon the acceptance of this work at https://github.com/BioMedIA-MBZUAI/DynaMMo.
Envisioning MedCLIP: A Deep Dive into Explainability for Medical Vision-Language Models
Mar 27, 2024Abstract:Explaining Deep Learning models is becoming increasingly important in the face of daily emerging multimodal models, particularly in safety-critical domains like medical imaging. However, the lack of detailed investigations into the performance of explainability methods on these models is widening the gap between their development and safe deployment. In this work, we analyze the performance of various explainable AI methods on a vision-language model, MedCLIP, to demystify its inner workings. We also provide a simple methodology to overcome the shortcomings of these methods. Our work offers a different new perspective on the explainability of a recent well-known VLM in the medical domain and our assessment method is generalizable to other current and possible future VLMs.
FissionFusion: Fast Geometric Generation and Hierarchical Souping for Medical Image Analysis
Mar 20, 2024



Abstract:The scarcity of well-annotated medical datasets requires leveraging transfer learning from broader datasets like ImageNet or pre-trained models like CLIP. Model soups averages multiple fine-tuned models aiming to improve performance on In-Domain (ID) tasks and enhance robustness against Out-of-Distribution (OOD) datasets. However, applying these methods to the medical imaging domain faces challenges and results in suboptimal performance. This is primarily due to differences in error surface characteristics that stem from data complexities such as heterogeneity, domain shift, class imbalance, and distributional shifts between training and testing phases. To address this issue, we propose a hierarchical merging approach that involves local and global aggregation of models at various levels based on models' hyperparameter configurations. Furthermore, to alleviate the need for training a large number of models in the hyperparameter search, we introduce a computationally efficient method using a cyclical learning rate scheduler to produce multiple models for aggregation in the weight space. Our method demonstrates significant improvements over the model souping approach across multiple datasets (around 6% gain in HAM10000 and CheXpert datasets) while maintaining low computational costs for model generation and selection. Moreover, we achieve better results on OOD datasets than model soups. The code is available at https://github.com/BioMedIA-MBZUAI/FissionFusion.
MedMerge: Merging Models for Effective Transfer Learning to Medical Imaging Tasks
Mar 18, 2024



Abstract:Transfer learning has become a powerful tool to initialize deep learning models to achieve faster convergence and higher performance. This is especially useful in the medical imaging analysis domain, where data scarcity limits possible performance gains for deep learning models. Some advancements have been made in boosting the transfer learning performance gain by merging models starting from the same initialization. However, in the medical imaging analysis domain, there is an opportunity in merging models starting from different initialisations, thus combining the features learnt from different tasks. In this work, we propose MedMerge, a method whereby the weights of different models can be merged, and their features can be effectively utilized to boost performance on a new task. With MedMerge, we learn kernel-level weights that can later be used to merge the models into a single model, even when starting from different initializations. Testing on various medical imaging analysis tasks, we show that our merged model can achieve significant performance gains, with up to 3% improvement on the F1 score. The code implementation of this work will be available at www.github.com/BioMedIA-MBZUAI/MedMerge.
Advanced Tumor Segmentation in Medical Imaging: An Ensemble Approach for BraTS 2023 Adult Glioma and Pediatric Tumor Tasks
Mar 14, 2024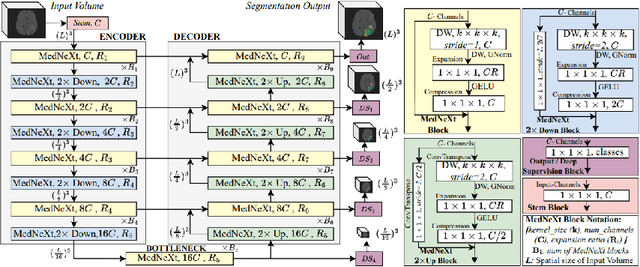
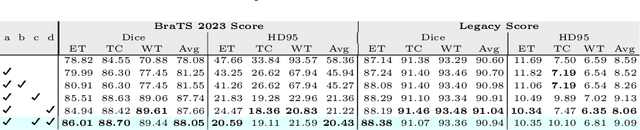
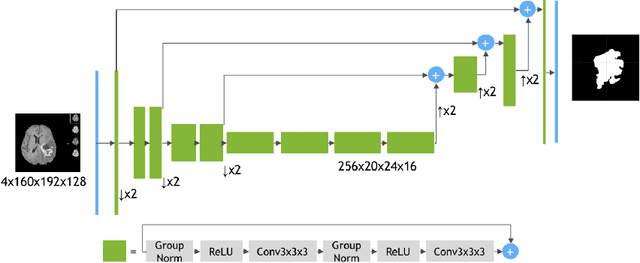
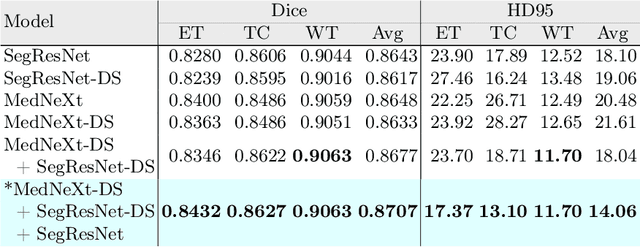
Abstract:Automated segmentation proves to be a valuable tool in precisely detecting tumors within medical images. The accurate identification and segmentation of tumor types hold paramount importance in diagnosing, monitoring, and treating highly fatal brain tumors. The BraTS challenge serves as a platform for researchers to tackle this issue by participating in open challenges focused on tumor segmentation. This study outlines our methodology for segmenting tumors in the context of two distinct tasks from the BraTS 2023 challenge: Adult Glioma and Pediatric Tumors. Our approach leverages two encoder-decoder-based CNN models, namely SegResNet and MedNeXt, for segmenting three distinct subregions of tumors. We further introduce a set of robust postprocessing to improve the segmentation, especially for the newly introduced BraTS 2023 metrics. The specifics of our approach and comprehensive performance analyses are expounded upon in this work. Our proposed approach achieves third place in the BraTS 2023 Adult Glioma Segmentation Challenges with an average of 0.8313 and 36.38 Dice and HD95 scores on the test set, respectively.
XReal: Realistic Anatomy and Pathology-Aware X-ray Generation via Controllable Diffusion Model
Mar 14, 2024



Abstract:Large-scale generative models have demonstrated impressive capacity in producing visually compelling images, with increasing applications in medical imaging. However, they continue to grapple with the challenge of image hallucination and the generation of anatomically inaccurate outputs. These limitations are mainly due to the sole reliance on textual inputs and lack of spatial control over the generated images, hindering the potential usefulness of such models in real-life settings. We present XReal, a novel controllable diffusion model for generating realistic chest X-ray images through precise anatomy and pathology location control. Our lightweight method can seamlessly integrate spatial control in a pre-trained text-to-image diffusion model without fine-tuning, retaining its existing knowledge while enhancing its generation capabilities. XReal outperforms state-of-the-art x-ray diffusion models in quantitative and qualitative metrics while showing 13% and 10% anatomy and pathology realism gain, respectively, based on the expert radiologist evaluation. Our model holds promise for advancing generative models in medical imaging, offering greater precision and adaptability while inviting further exploration in this evolving field. A large synthetically generated data with annotations and code is publicly available at https://github.com/BioMedIA-MBZUAI/XReal.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge