Fadillah Maani
FetalCLIP: A Visual-Language Foundation Model for Fetal Ultrasound Image Analysis
Feb 20, 2025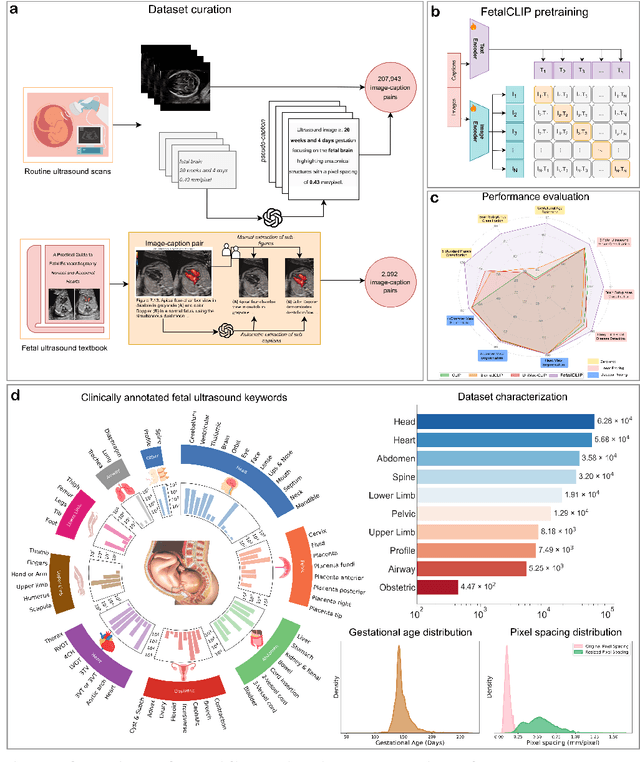
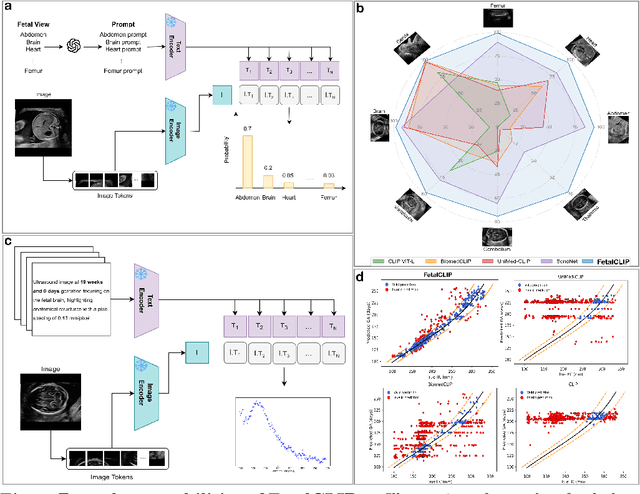
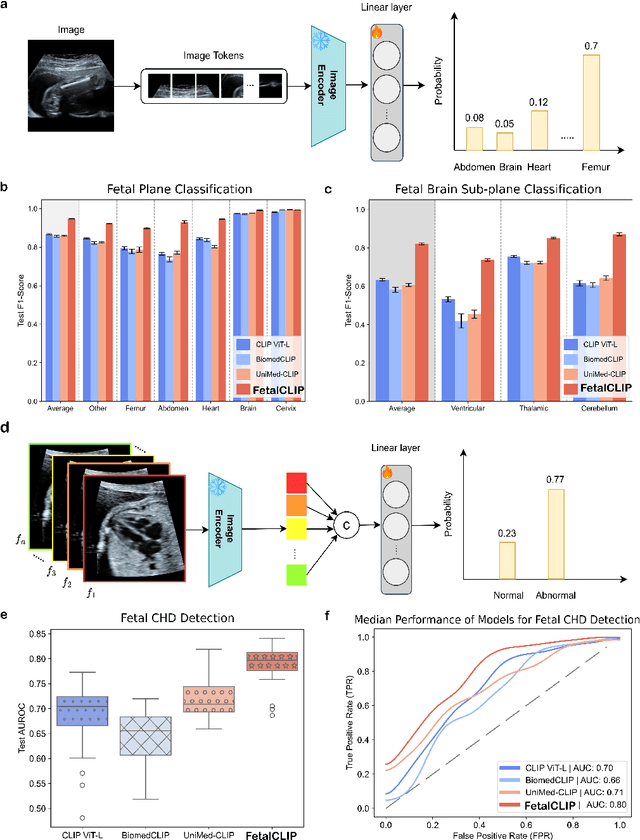
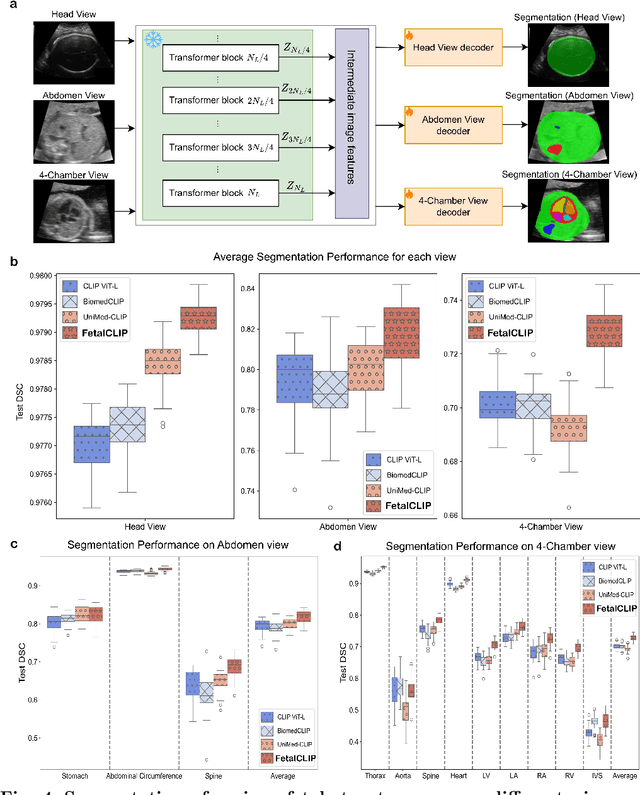
Abstract:Foundation models are becoming increasingly effective in the medical domain, offering pre-trained models on large datasets that can be readily adapted for downstream tasks. Despite progress, fetal ultrasound images remain a challenging domain for foundation models due to their inherent complexity, often requiring substantial additional training and facing limitations due to the scarcity of paired multimodal data. To overcome these challenges, here we introduce FetalCLIP, a vision-language foundation model capable of generating universal representation of fetal ultrasound images. FetalCLIP was pre-trained using a multimodal learning approach on a diverse dataset of 210,035 fetal ultrasound images paired with text. This represents the largest paired dataset of its kind used for foundation model development to date. This unique training approach allows FetalCLIP to effectively learn the intricate anatomical features present in fetal ultrasound images, resulting in robust representations that can be used for a variety of downstream applications. In extensive benchmarking across a range of key fetal ultrasound applications, including classification, gestational age estimation, congenital heart defect (CHD) detection, and fetal structure segmentation, FetalCLIP outperformed all baselines while demonstrating remarkable generalizability and strong performance even with limited labeled data. We plan to release the FetalCLIP model publicly for the benefit of the broader scientific community.
On Enhancing Brain Tumor Segmentation Across Diverse Populations with Convolutional Neural Networks
May 05, 2024

Abstract:Brain tumor segmentation is a fundamental step in assessing a patient's cancer progression. However, manual segmentation demands significant expert time to identify tumors in 3D multimodal brain MRI scans accurately. This reliance on manual segmentation makes the process prone to intra- and inter-observer variability. This work proposes a brain tumor segmentation method as part of the BraTS-GoAT challenge. The task is to segment tumors in brain MRI scans automatically from various populations, such as adults, pediatrics, and underserved sub-Saharan Africa. We employ a recent CNN architecture for medical image segmentation, namely MedNeXt, as our baseline, and we implement extensive model ensembling and postprocessing for inference. Our experiments show that our method performs well on the unseen validation set with an average DSC of 85.54% and HD95 of 27.88. The code is available on https://github.com/BioMedIA-MBZUAI/BraTS2024_BioMedIAMBZ.
Advanced Tumor Segmentation in Medical Imaging: An Ensemble Approach for BraTS 2023 Adult Glioma and Pediatric Tumor Tasks
Mar 14, 2024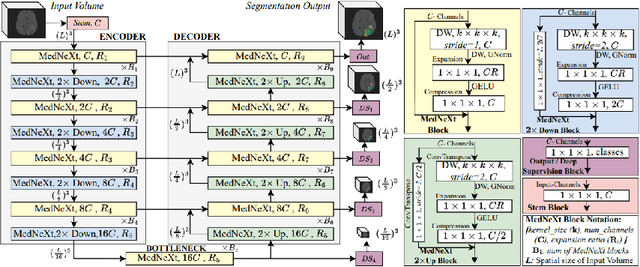
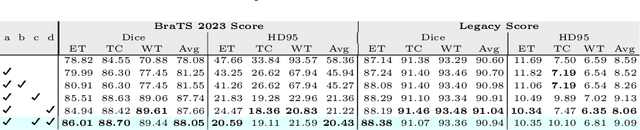
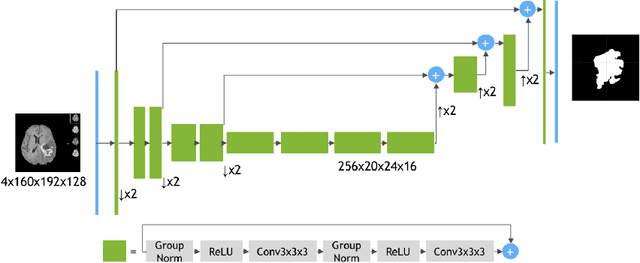
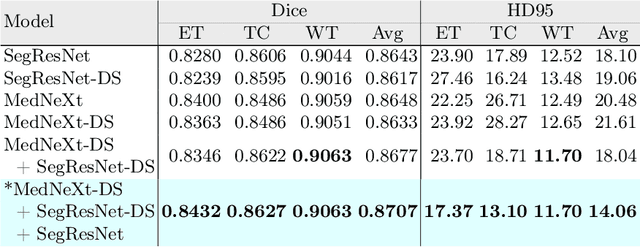
Abstract:Automated segmentation proves to be a valuable tool in precisely detecting tumors within medical images. The accurate identification and segmentation of tumor types hold paramount importance in diagnosing, monitoring, and treating highly fatal brain tumors. The BraTS challenge serves as a platform for researchers to tackle this issue by participating in open challenges focused on tumor segmentation. This study outlines our methodology for segmenting tumors in the context of two distinct tasks from the BraTS 2023 challenge: Adult Glioma and Pediatric Tumors. Our approach leverages two encoder-decoder-based CNN models, namely SegResNet and MedNeXt, for segmenting three distinct subregions of tumors. We further introduce a set of robust postprocessing to improve the segmentation, especially for the newly introduced BraTS 2023 metrics. The specifics of our approach and comprehensive performance analyses are expounded upon in this work. Our proposed approach achieves third place in the BraTS 2023 Adult Glioma Segmentation Challenges with an average of 0.8313 and 36.38 Dice and HD95 scores on the test set, respectively.
UniLVSeg: Unified Left Ventricular Segmentation with Sparsely Annotated Echocardiogram Videos through Self-Supervised Temporal Masking and Weakly Supervised Training
Sep 30, 2023Abstract:Echocardiography has become an indispensable clinical imaging modality for general heart health assessment. From calculating biomarkers such as ejection fraction to the probability of a patient's heart failure, accurate segmentation of the heart and its structures allows doctors to plan and execute treatments with greater precision and accuracy. However, achieving accurate and robust left ventricle segmentation is time-consuming and challenging due to different reasons. This work introduces a novel approach for consistent left ventricular (LV) segmentation from sparsely annotated echocardiogram videos. We achieve this through (1) self-supervised learning (SSL) using temporal masking followed by (2) weakly supervised training. We investigate two different segmentation approaches: 3D segmentation and a novel 2D superimage (SI). We demonstrate how our proposed method outperforms the state-of-the-art solutions by achieving a 93.32% (95%CI 93.21-93.43%) dice score on a large-scale dataset (EchoNet-Dynamic) while being more efficient. To show the effectiveness of our approach, we provide extensive ablation studies, including pre-training settings and various deep learning backbones. Additionally, we discuss how our proposed methodology achieves high data utility by incorporating unlabeled frames in the training process. To help support the AI in medicine community, the complete solution with the source code will be made publicly available upon acceptance.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge