Numan Saeed
AnatomiX, an Anatomy-Aware Grounded Multimodal Large Language Model for Chest X-Ray Interpretation
Jan 06, 2026Abstract:Multimodal medical large language models have shown impressive progress in chest X-ray interpretation but continue to face challenges in spatial reasoning and anatomical understanding. Although existing grounding techniques improve overall performance, they often fail to establish a true anatomical correspondence, resulting in incorrect anatomical understanding in the medical domain. To address this gap, we introduce AnatomiX, a multitask multimodal large language model explicitly designed for anatomically grounded chest X-ray interpretation. Inspired by the radiological workflow, AnatomiX adopts a two stage approach: first, it identifies anatomical structures and extracts their features, and then leverages a large language model to perform diverse downstream tasks such as phrase grounding, report generation, visual question answering, and image understanding. Extensive experiments across multiple benchmarks demonstrate that AnatomiX achieves superior anatomical reasoning and delivers over 25% improvement in performance on anatomy grounding, phrase grounding, grounded diagnosis and grounded captioning tasks compared to existing approaches. Code and pretrained model are available at https://github.com/aneesurhashmi/anatomix
FETAL-GAUGE: A Benchmark for Assessing Vision-Language Models in Fetal Ultrasound
Dec 25, 2025Abstract:The growing demand for prenatal ultrasound imaging has intensified a global shortage of trained sonographers, creating barriers to essential fetal health monitoring. Deep learning has the potential to enhance sonographers' efficiency and support the training of new practitioners. Vision-Language Models (VLMs) are particularly promising for ultrasound interpretation, as they can jointly process images and text to perform multiple clinical tasks within a single framework. However, despite the expansion of VLMs, no standardized benchmark exists to evaluate their performance in fetal ultrasound imaging. This gap is primarily due to the modality's challenging nature, operator dependency, and the limited public availability of datasets. To address this gap, we present Fetal-Gauge, the first and largest visual question answering benchmark specifically designed to evaluate VLMs across various fetal ultrasound tasks. Our benchmark comprises over 42,000 images and 93,000 question-answer pairs, spanning anatomical plane identification, visual grounding of anatomical structures, fetal orientation assessment, clinical view conformity, and clinical diagnosis. We systematically evaluate several state-of-the-art VLMs, including general-purpose and medical-specific models, and reveal a substantial performance gap: the best-performing model achieves only 55\% accuracy, far below clinical requirements. Our analysis identifies critical limitations of current VLMs in fetal ultrasound interpretation, highlighting the urgent need for domain-adapted architectures and specialized training approaches. Fetal-Gauge establishes a rigorous foundation for advancing multimodal deep learning in prenatal care and provides a pathway toward addressing global healthcare accessibility challenges. Our benchmark will be publicly available once the paper gets accepted.
MAFM^3: Modular Adaptation of Foundation Models for Multi-Modal Medical AI
Nov 14, 2025Abstract:Foundational models are trained on extensive datasets to capture the general trends of a domain. However, in medical imaging, the scarcity of data makes pre-training for every domain, modality, or task challenging. Instead of building separate models, we propose MAFM^3 (Modular Adaptation of Foundation Models for Multi-Modal Medical AI), a framework that enables a single foundation model to expand into diverse domains, tasks, and modalities through lightweight modular components. These components serve as specialized skill sets that allow the system to flexibly activate the appropriate capability at the inference time, depending on the input type or clinical objective. Unlike conventional adaptation methods that treat each new task or modality in isolation, MAFM^3 provides a unified and expandable framework for efficient multitask and multimodality adaptation. Empirically, we validate our approach by adapting a chest CT foundation model initially trained for classification into prognosis and segmentation modules. Our results show improved performance on both tasks. Furthermore, by incorporating PET scans, MAFM^3 achieved an improvement in the Dice score 5% compared to the respective baselines. These findings establish that foundation models, when equipped with modular components, are not inherently constrained to their initial training scope but can evolve into multitask, multimodality systems for medical imaging. The code implementation of this work can be found at https://github.com/Areeb2735/CTscan_prognosis_VLM
CardioBench: Do Echocardiography Foundation Models Generalize Beyond the Lab?
Oct 01, 2025Abstract:Foundation models (FMs) are reshaping medical imaging, yet their application in echocardiography remains limited. While several echocardiography-specific FMs have recently been introduced, no standardized benchmark exists to evaluate them. Echocardiography poses unique challenges, including noisy acquisitions, high frame redundancy, and limited public datasets. Most existing solutions evaluate on private data, restricting comparability. To address this, we introduce CardioBench, a comprehensive benchmark for echocardiography FMs. CardioBench unifies eight publicly available datasets into a standardized suite spanning four regression and five classification tasks, covering functional, structural, diagnostic, and view recognition endpoints. We evaluate several leading FM, including cardiac-specific, biomedical, and general-purpose encoders, under consistent zero-shot, probing, and alignment protocols. Our results highlight complementary strengths across model families: temporal modeling is critical for functional regression, retrieval provides robustness under distribution shift, and domain-specific text encoders capture physiologically meaningful axes. General-purpose encoders transfer strongly and often close the gap with probing, but struggle with fine-grained distinctions like view classification and subtle pathology recognition. By releasing preprocessing, splits, and public evaluation pipelines, CardioBench establishes a reproducible reference point and offers actionable insights to guide the design of future echocardiography foundation models.
PCA-Guided Autoencoding for Structured Dimensionality Reduction in Active Infrared Thermography
Aug 11, 2025Abstract:Active Infrared thermography (AIRT) is a widely adopted non-destructive testing (NDT) technique for detecting subsurface anomalies in industrial components. Due to the high dimensionality of AIRT data, current approaches employ non-linear autoencoders (AEs) for dimensionality reduction. However, the latent space learned by AIRT AEs lacks structure, limiting their effectiveness in downstream defect characterization tasks. To address this limitation, this paper proposes a principal component analysis guided (PCA-guided) autoencoding framework for structured dimensionality reduction to capture intricate, non-linear features in thermographic signals while enforcing a structured latent space. A novel loss function, PCA distillation loss, is introduced to guide AIRT AEs to align the latent representation with structured PCA components while capturing the intricate, non-linear patterns in thermographic signals. To evaluate the utility of the learned, structured latent space, we propose a neural network-based evaluation metric that assesses its suitability for defect characterization. Experimental results show that the proposed PCA-guided AE outperforms state-of-the-art dimensionality reduction methods on PVC, CFRP, and PLA samples in terms of contrast, signal-to-noise ratio (SNR), and neural network-based metrics.
Efficient Parameter Adaptation for Multi-Modal Medical Image Segmentation and Prognosis
Apr 18, 2025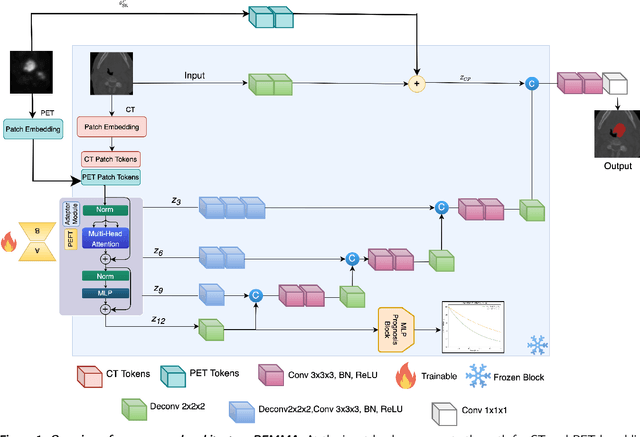
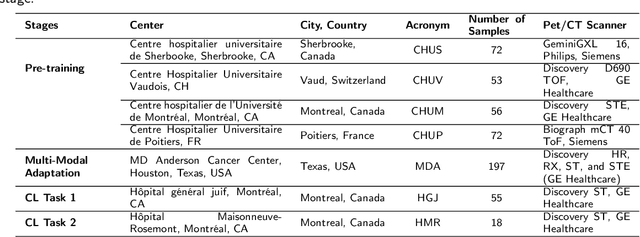
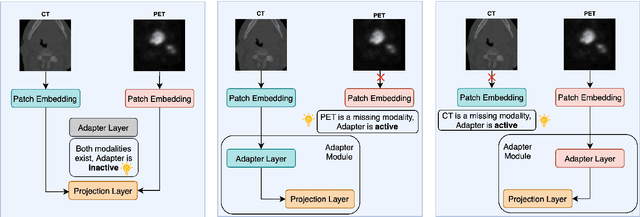
Abstract:Cancer detection and prognosis relies heavily on medical imaging, particularly CT and PET scans. Deep Neural Networks (DNNs) have shown promise in tumor segmentation by fusing information from these modalities. However, a critical bottleneck exists: the dependency on CT-PET data concurrently for training and inference, posing a challenge due to the limited availability of PET scans. Hence, there is a clear need for a flexible and efficient framework that can be trained with the widely available CT scans and can be still adapted for PET scans when they become available. In this work, we propose a parameter-efficient multi-modal adaptation (PEMMA) framework for lightweight upgrading of a transformer-based segmentation model trained only on CT scans such that it can be efficiently adapted for use with PET scans when they become available. This framework is further extended to perform prognosis task maintaining the same efficient cross-modal fine-tuning approach. The proposed approach is tested with two well-known segementation backbones, namely UNETR and Swin UNETR. Our approach offers two main advantages. Firstly, we leverage the inherent modularity of the transformer architecture and perform low-rank adaptation (LoRA) as well as decomposed low-rank adaptation (DoRA) of the attention weights to achieve parameter-efficient adaptation. Secondly, by minimizing cross-modal entanglement, PEMMA allows updates using only one modality without causing catastrophic forgetting in the other. Our method achieves comparable performance to early fusion, but with only 8% of the trainable parameters, and demonstrates a significant +28% Dice score improvement on PET scans when trained with a single modality. Furthermore, in prognosis, our method improves the concordance index by +10% when adapting a CT-pretrained model to include PET scans, and by +23% when adapting for both PET and EHR data.
In-Model Merging for Enhancing the Robustness of Medical Imaging Classification Models
Feb 27, 2025



Abstract:Model merging is an effective strategy to merge multiple models for enhancing model performances, and more efficient than ensemble learning as it will not introduce extra computation into inference. However, limited research explores if the merging process can occur within one model and enhance the model's robustness, which is particularly critical in the medical image domain. In the paper, we are the first to propose in-model merging (InMerge), a novel approach that enhances the model's robustness by selectively merging similar convolutional kernels in the deep layers of a single convolutional neural network (CNN) during the training process for classification. We also analytically reveal important characteristics that affect how in-model merging should be performed, serving as an insightful reference for the community. We demonstrate the feasibility and effectiveness of this technique for different CNN architectures on 4 prevalent datasets. The proposed InMerge-trained model surpasses the typically-trained model by a substantial margin. The code will be made public.
FetalCLIP: A Visual-Language Foundation Model for Fetal Ultrasound Image Analysis
Feb 20, 2025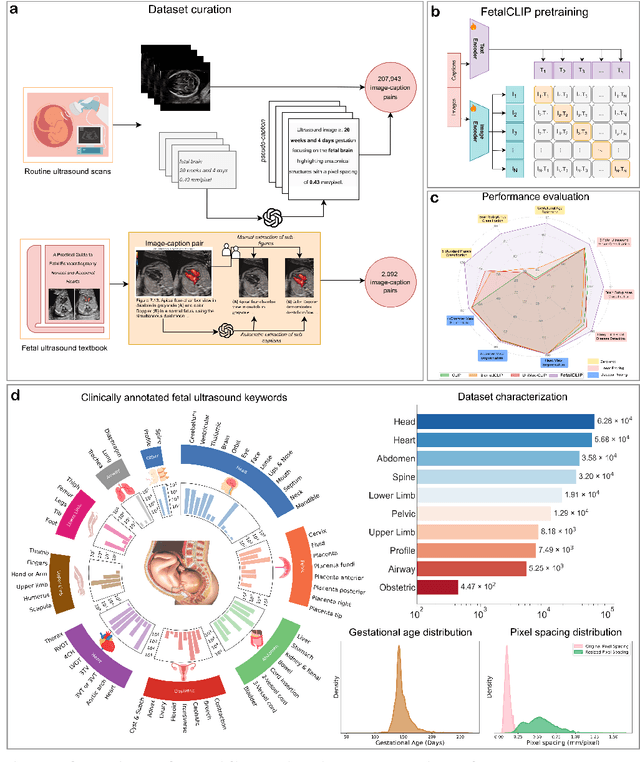
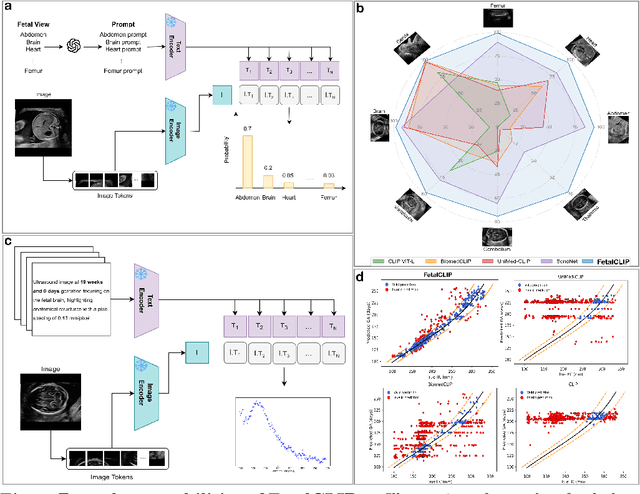
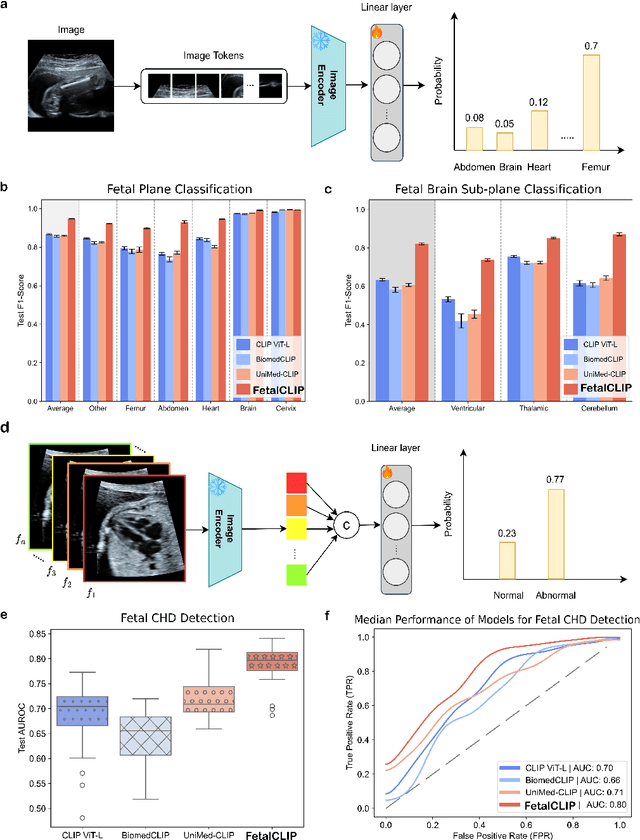
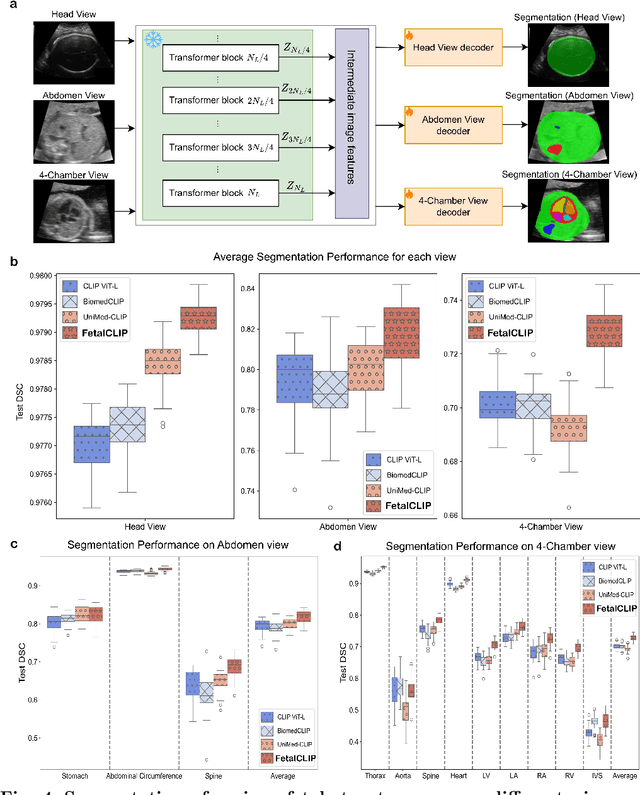
Abstract:Foundation models are becoming increasingly effective in the medical domain, offering pre-trained models on large datasets that can be readily adapted for downstream tasks. Despite progress, fetal ultrasound images remain a challenging domain for foundation models due to their inherent complexity, often requiring substantial additional training and facing limitations due to the scarcity of paired multimodal data. To overcome these challenges, here we introduce FetalCLIP, a vision-language foundation model capable of generating universal representation of fetal ultrasound images. FetalCLIP was pre-trained using a multimodal learning approach on a diverse dataset of 210,035 fetal ultrasound images paired with text. This represents the largest paired dataset of its kind used for foundation model development to date. This unique training approach allows FetalCLIP to effectively learn the intricate anatomical features present in fetal ultrasound images, resulting in robust representations that can be used for a variety of downstream applications. In extensive benchmarking across a range of key fetal ultrasound applications, including classification, gestational age estimation, congenital heart defect (CHD) detection, and fetal structure segmentation, FetalCLIP outperformed all baselines while demonstrating remarkable generalizability and strong performance even with limited labeled data. We plan to release the FetalCLIP model publicly for the benefit of the broader scientific community.
SurvCORN: Survival Analysis with Conditional Ordinal Ranking Neural Network
Sep 30, 2024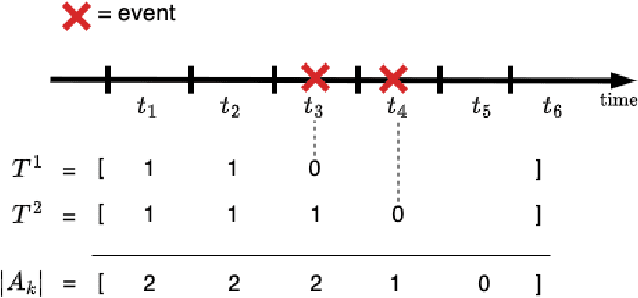
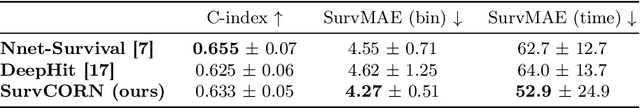
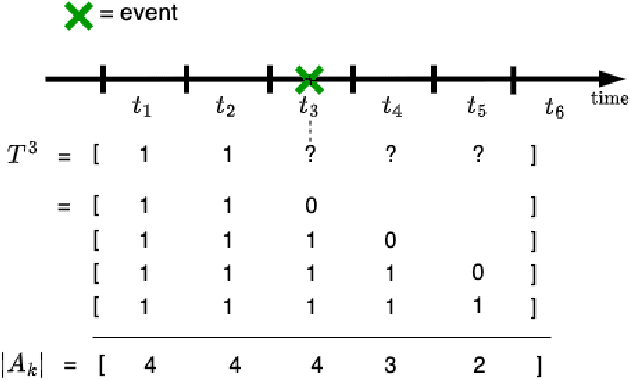
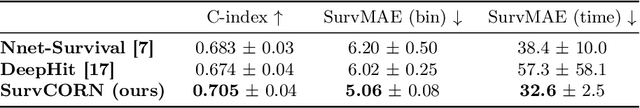
Abstract:Survival analysis plays a crucial role in estimating the likelihood of future events for patients by modeling time-to-event data, particularly in healthcare settings where predictions about outcomes such as death and disease recurrence are essential. However, this analysis poses challenges due to the presence of censored data, where time-to-event information is missing for certain data points. Yet, censored data can offer valuable insights, provided we appropriately incorporate the censoring time during modeling. In this paper, we propose SurvCORN, a novel method utilizing conditional ordinal ranking networks to predict survival curves directly. Additionally, we introduce SurvMAE, a metric designed to evaluate the accuracy of model predictions in estimating time-to-event outcomes. Through empirical evaluation on two real-world cancer datasets, we demonstrate SurvCORN's ability to maintain accurate ordering between patient outcomes while improving individual time-to-event predictions. Our contributions extend recent advancements in ordinal regression to survival analysis, offering valuable insights into accurate prognosis in healthcare settings.
On Evaluating Adversarial Robustness of Volumetric Medical Segmentation Models
Jun 12, 2024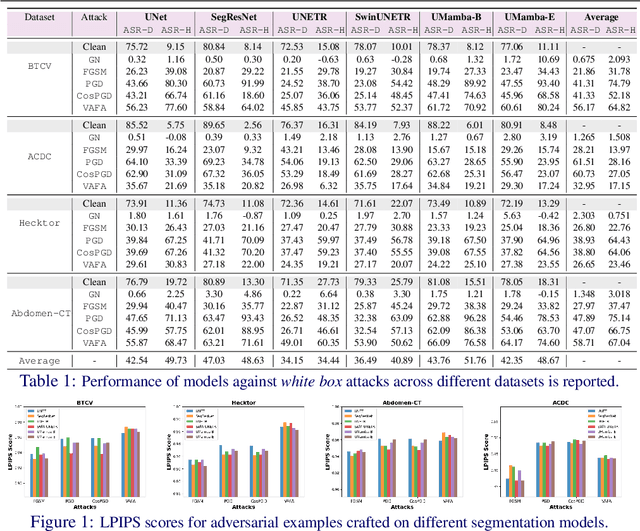
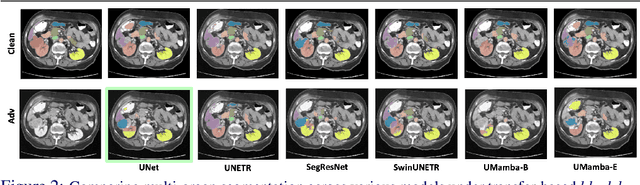
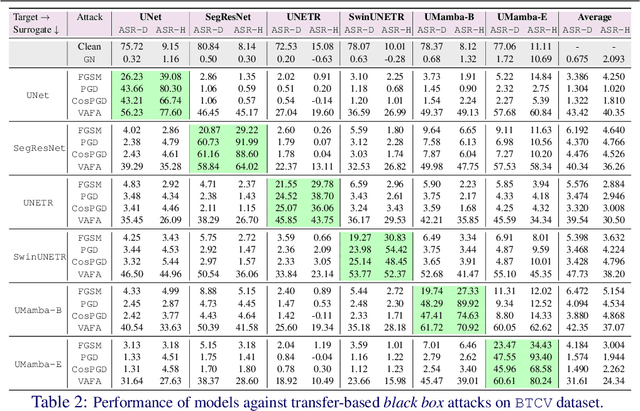
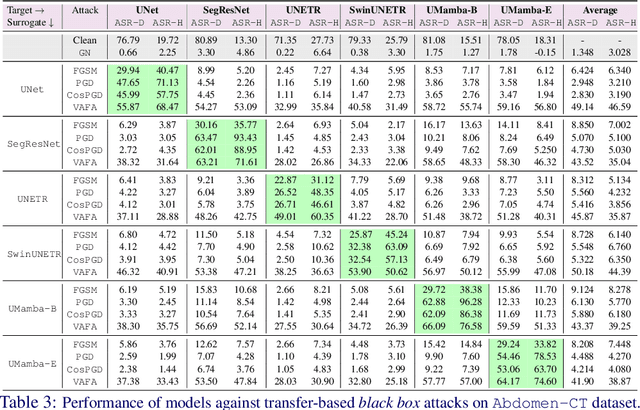
Abstract:Volumetric medical segmentation models have achieved significant success on organ and tumor-based segmentation tasks in recent years. However, their vulnerability to adversarial attacks remains largely unexplored, raising serious concerns regarding the real-world deployment of tools employing such models in the healthcare sector. This underscores the importance of investigating the robustness of existing models. In this context, our work aims to empirically examine the adversarial robustness across current volumetric segmentation architectures, encompassing Convolutional, Transformer, and Mamba-based models. We extend this investigation across four volumetric segmentation datasets, evaluating robustness under both white box and black box adversarial attacks. Overall, we observe that while both pixel and frequency-based attacks perform reasonably well under white box setting, the latter performs significantly better under transfer-based black box attacks. Across our experiments, we observe transformer-based models show higher robustness than convolution-based models with Mamba-based models being the most vulnerable. Additionally, we show that large-scale training of volumetric segmentation models improves the model's robustness against adversarial attacks. The code and pretrained models will be made available at https://github.com/HashmatShadab/Robustness-of-Volumetric-Medical-Segmentation-Models.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge