Kudaibergen Abutalip
EDUE: Expert Disagreement-Guided One-Pass Uncertainty Estimation for Medical Image Segmentation
Mar 25, 2024
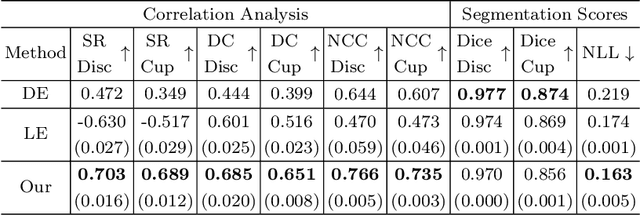
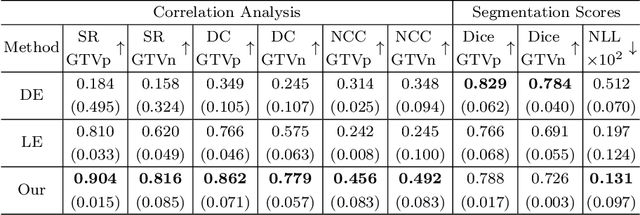

Abstract:Deploying deep learning (DL) models in medical applications relies on predictive performance and other critical factors, such as conveying trustworthy predictive uncertainty. Uncertainty estimation (UE) methods provide potential solutions for evaluating prediction reliability and improving the model confidence calibration. Despite increasing interest in UE, challenges persist, such as the need for explicit methods to capture aleatoric uncertainty and align uncertainty estimates with real-life disagreements among domain experts. This paper proposes an Expert Disagreement-Guided Uncertainty Estimation (EDUE) for medical image segmentation. By leveraging variability in ground-truth annotations from multiple raters, we guide the model during training and incorporate random sampling-based strategies to enhance calibration confidence. Our method achieves 55% and 23% improvement in correlation on average with expert disagreements at the image and pixel levels, respectively, better calibration, and competitive segmentation performance compared to the state-of-the-art deep ensembles, requiring only a single forward pass.
Improving Stain Invariance of CNNs for Segmentation by Fusing Channel Attention and Domain-Adversarial Training
Apr 22, 2023
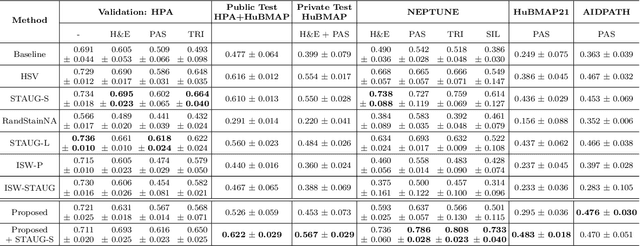
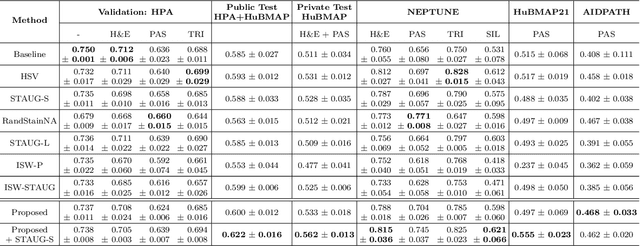
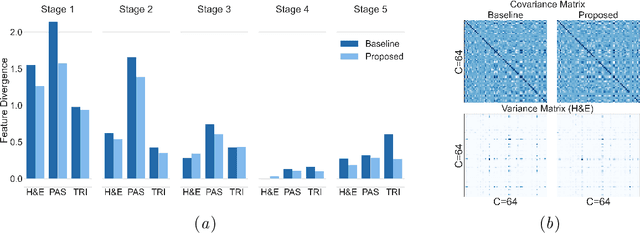
Abstract:Variability in staining protocols, such as different slide preparation techniques, chemicals, and scanner configurations, can result in a diverse set of whole slide images (WSIs). This distribution shift can negatively impact the performance of deep learning models on unseen samples, presenting a significant challenge for developing new computational pathology applications. In this study, we propose a method for improving the generalizability of convolutional neural networks (CNNs) to stain changes in a single-source setting for semantic segmentation. Recent studies indicate that style features mainly exist as covariances in earlier network layers. We design a channel attention mechanism based on these findings that detects stain-specific features and modify the previously proposed stain-invariant training scheme. We reweigh the outputs of earlier layers and pass them to the stain-adversarial training branch. We evaluate our method on multi-center, multi-stain datasets and demonstrate its effectiveness through interpretability analysis. Our approach achieves substantial improvements over baselines and competitive performance compared to other methods, as measured by various evaluation metrics. We also show that combining our method with stain augmentation leads to mutually beneficial results and outperforms other techniques. Overall, our study makes significant contributions to the field of computational pathology.
Is it Possible to Predict MGMT Promoter Methylation from Brain Tumor MRI Scans using Deep Learning Models?
Jan 16, 2022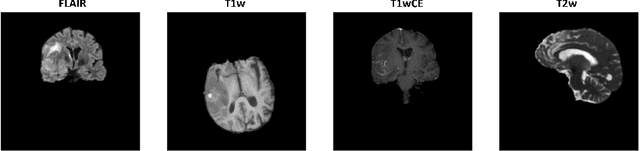
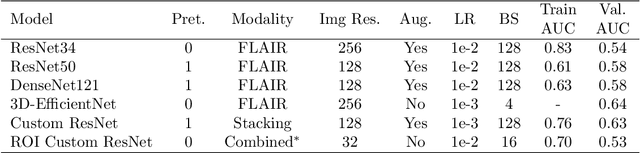
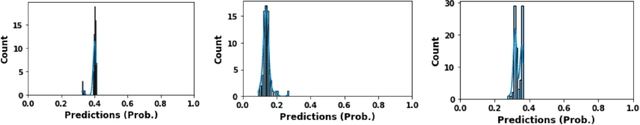

Abstract:Glioblastoma is a common brain malignancy that tends to occur in older adults and is almost always lethal. The effectiveness of chemotherapy, being the standard treatment for most cancer types, can be improved if a particular genetic sequence in the tumor known as MGMT promoter is methylated. However, to identify the state of the MGMT promoter, the conventional approach is to perform a biopsy for genetic analysis, which is time and effort consuming. A couple of recent publications proposed a connection between the MGMT promoter state and the MRI scans of the tumor and hence suggested the use of deep learning models for this purpose. Therefore, in this work, we use one of the most extensive datasets, BraTS 2021, to study the potency of employing deep learning solutions, including 2D and 3D CNN models and vision transformers. After conducting a thorough analysis of the models' performance, we concluded that there seems to be no connection between the MRI scans and the state of the MGMT promoter.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge