Ikboljon Sobirov
EDUE: Expert Disagreement-Guided One-Pass Uncertainty Estimation for Medical Image Segmentation
Mar 25, 2024
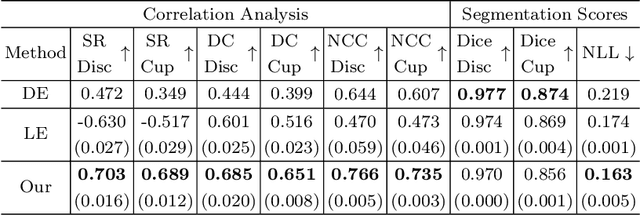
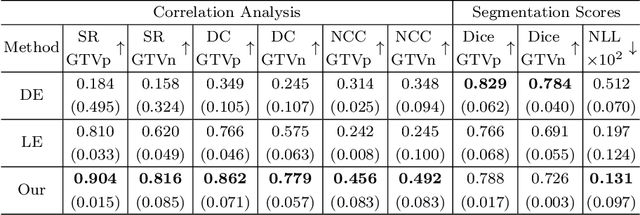

Abstract:Deploying deep learning (DL) models in medical applications relies on predictive performance and other critical factors, such as conveying trustworthy predictive uncertainty. Uncertainty estimation (UE) methods provide potential solutions for evaluating prediction reliability and improving the model confidence calibration. Despite increasing interest in UE, challenges persist, such as the need for explicit methods to capture aleatoric uncertainty and align uncertainty estimates with real-life disagreements among domain experts. This paper proposes an Expert Disagreement-Guided Uncertainty Estimation (EDUE) for medical image segmentation. By leveraging variability in ground-truth annotations from multiple raters, we guide the model during training and incorporate random sampling-based strategies to enhance calibration confidence. Our method achieves 55% and 23% improvement in correlation on average with expert disagreements at the image and pixel levels, respectively, better calibration, and competitive segmentation performance compared to the state-of-the-art deep ensembles, requiring only a single forward pass.
HuLP: Human-in-the-Loop for Prognosis
Mar 19, 2024Abstract:This paper introduces HuLP, a Human-in-the-Loop for Prognosis model designed to enhance the reliability and interpretability of prognostic models in clinical contexts, especially when faced with the complexities of missing covariates and outcomes. HuLP offers an innovative approach that enables human expert intervention, empowering clinicians to interact with and correct models' predictions, thus fostering collaboration between humans and AI models to produce more accurate prognosis. Additionally, HuLP addresses the challenges of missing data by utilizing neural networks and providing a tailored methodology that effectively handles missing data. Traditional methods often struggle to capture the nuanced variations within patient populations, leading to compromised prognostic predictions. HuLP imputes missing covariates based on imaging features, aligning more closely with clinician workflows and enhancing reliability. We conduct our experiments on two real-world, publicly available medical datasets to demonstrate the superiority of HuLP.
Advanced Tumor Segmentation in Medical Imaging: An Ensemble Approach for BraTS 2023 Adult Glioma and Pediatric Tumor Tasks
Mar 14, 2024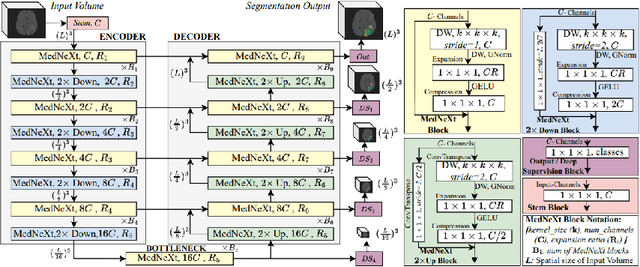
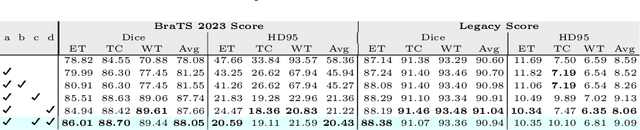
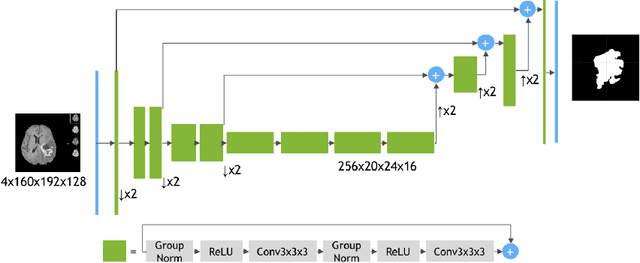
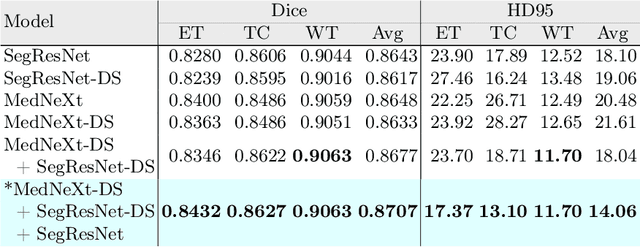
Abstract:Automated segmentation proves to be a valuable tool in precisely detecting tumors within medical images. The accurate identification and segmentation of tumor types hold paramount importance in diagnosing, monitoring, and treating highly fatal brain tumors. The BraTS challenge serves as a platform for researchers to tackle this issue by participating in open challenges focused on tumor segmentation. This study outlines our methodology for segmenting tumors in the context of two distinct tasks from the BraTS 2023 challenge: Adult Glioma and Pediatric Tumors. Our approach leverages two encoder-decoder-based CNN models, namely SegResNet and MedNeXt, for segmenting three distinct subregions of tumors. We further introduce a set of robust postprocessing to improve the segmentation, especially for the newly introduced BraTS 2023 metrics. The specifics of our approach and comprehensive performance analyses are expounded upon in this work. Our proposed approach achieves third place in the BraTS 2023 Adult Glioma Segmentation Challenges with an average of 0.8313 and 36.38 Dice and HD95 scores on the test set, respectively.
Reinforcement Learning for Solving Stochastic Vehicle Routing Problem with Time Windows
Feb 15, 2024Abstract:This paper introduces a reinforcement learning approach to optimize the Stochastic Vehicle Routing Problem with Time Windows (SVRP), focusing on reducing travel costs in goods delivery. We develop a novel SVRP formulation that accounts for uncertain travel costs and demands, alongside specific customer time windows. An attention-based neural network trained through reinforcement learning is employed to minimize routing costs. Our approach addresses a gap in SVRP research, which traditionally relies on heuristic methods, by leveraging machine learning. The model outperforms the Ant-Colony Optimization algorithm, achieving a 1.73% reduction in travel costs. It uniquely integrates external information, demonstrating robustness in diverse environments, making it a valuable benchmark for future SVRP studies and industry application.
Reinforcement Learning for Solving Stochastic Vehicle Routing Problem
Nov 13, 2023Abstract:This study addresses a gap in the utilization of Reinforcement Learning (RL) and Machine Learning (ML) techniques in solving the Stochastic Vehicle Routing Problem (SVRP) that involves the challenging task of optimizing vehicle routes under uncertain conditions. We propose a novel end-to-end framework that comprehensively addresses the key sources of stochasticity in SVRP and utilizes an RL agent with a simple yet effective architecture and a tailored training method. Through comparative analysis, our proposed model demonstrates superior performance compared to a widely adopted state-of-the-art metaheuristic, achieving a significant 3.43% reduction in travel costs. Furthermore, the model exhibits robustness across diverse SVRP settings, highlighting its adaptability and ability to learn optimal routing strategies in varying environments. The publicly available implementation of our framework serves as a valuable resource for future research endeavors aimed at advancing RL-based solutions for SVRP.
LegoNet: Alternating Model Blocks for Medical Image Segmentation
Jun 06, 2023Abstract:Since the emergence of convolutional neural networks (CNNs), and later vision transformers (ViTs), the common paradigm for model development has always been using a set of identical block types with varying parameters/hyper-parameters. To leverage the benefits of different architectural designs (e.g. CNNs and ViTs), we propose to alternate structurally different types of blocks to generate a new architecture, mimicking how Lego blocks can be assembled together. Using two CNN-based and one SwinViT-based blocks, we investigate three variations to the so-called LegoNet that applies the new concept of block alternation for the segmentation task in medical imaging. We also study a new clinical problem which has not been investigated before, namely the right internal mammary artery (RIMA) and perivascular space segmentation from computed tomography angiography (CTA) which has demonstrated a prognostic value to major cardiovascular outcomes. We compare the model performance against popular CNN and ViT architectures using two large datasets (e.g. achieving 0.749 dice similarity coefficient (DSC) on the larger dataset). We evaluate the performance of the model on three external testing cohorts as well, where an expert clinician made corrections to the model segmented results (DSC>0.90 for the three cohorts). To assess our proposed model for suitability in clinical use, we perform intra- and inter-observer variability analysis. Finally, we investigate a joint self-supervised learning approach to assess its impact on model performance. The code and the pretrained model weights will be available upon acceptance.
Diagnosis and Prognosis of Head and Neck Cancer Patients using Artificial Intelligence
May 31, 2023



Abstract:Cancer is one of the most life-threatening diseases worldwide, and head and neck (H&N) cancer is a prevalent type with hundreds of thousands of new cases recorded each year. Clinicians use medical imaging modalities such as computed tomography and positron emission tomography to detect the presence of a tumor, and they combine that information with clinical data for patient prognosis. The process is mostly challenging and time-consuming. Machine learning and deep learning can automate these tasks to help clinicians with highly promising results. This work studies two approaches for H&N tumor segmentation: (i) exploration and comparison of vision transformer (ViT)-based and convolutional neural network-based models; and (ii) proposal of a novel 2D perspective to working with 3D data. Furthermore, this work proposes two new architectures for the prognosis task. An ensemble of several models predicts patient outcomes (which won the HECKTOR 2021 challenge prognosis task), and a ViT-based framework concurrently performs patient outcome prediction and tumor segmentation, which outperforms the ensemble model.
MGMT promoter methylation status prediction using MRI scans? An extensive experimental evaluation of deep learning models
Apr 03, 2023Abstract:The number of studies on deep learning for medical diagnosis is expanding, and these systems are often claimed to outperform clinicians. However, only a few systems have shown medical efficacy. From this perspective, we examine a wide range of deep learning algorithms for the assessment of glioblastoma - a common brain tumor in older adults that is lethal. Surgery, chemotherapy, and radiation are the standard treatments for glioblastoma patients. The methylation status of the MGMT promoter, a specific genetic sequence found in the tumor, affects chemotherapy's effectiveness. MGMT promoter methylation improves chemotherapy response and survival in several cancers. MGMT promoter methylation is determined by a tumor tissue biopsy, which is then genetically tested. This lengthy and invasive procedure increases the risk of infection and other complications. Thus, researchers have used deep learning models to examine the tumor from brain MRI scans to determine the MGMT promoter's methylation state. We employ deep learning models and one of the largest public MRI datasets of 585 participants to predict the methylation status of the MGMT promoter in glioblastoma tumors using MRI scans. We test these models using Grad-CAM, occlusion sensitivity, feature visualizations, and training loss landscapes. Our results show no correlation between these two, indicating that external cohort data should be used to verify these models' performance to assure the accuracy and reliability of deep learning systems in cancer diagnosis.
TMSS: An End-to-End Transformer-based Multimodal Network for Segmentation and Survival Prediction
Sep 12, 2022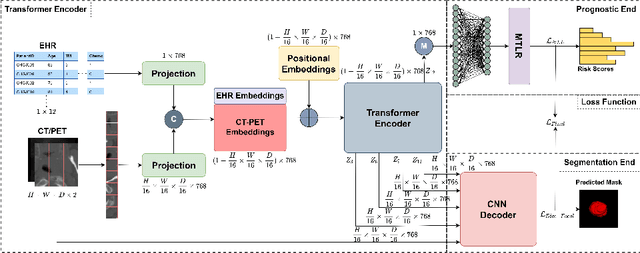

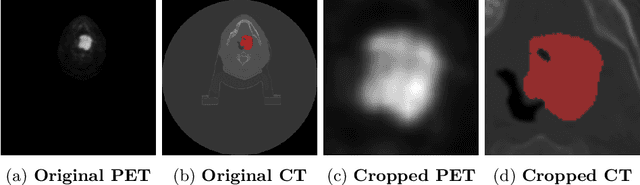
Abstract:When oncologists estimate cancer patient survival, they rely on multimodal data. Even though some multimodal deep learning methods have been proposed in the literature, the majority rely on having two or more independent networks that share knowledge at a later stage in the overall model. On the other hand, oncologists do not do this in their analysis but rather fuse the information in their brain from multiple sources such as medical images and patient history. This work proposes a deep learning method that mimics oncologists' analytical behavior when quantifying cancer and estimating patient survival. We propose TMSS, an end-to-end Transformer based Multimodal network for Segmentation and Survival prediction that leverages the superiority of transformers that lies in their abilities to handle different modalities. The model was trained and validated for segmentation and prognosis tasks on the training dataset from the HEad & NeCK TumOR segmentation and the outcome prediction in PET/CT images challenge (HECKTOR). We show that the proposed prognostic model significantly outperforms state-of-the-art methods with a concordance index of 0.763+/-0.14 while achieving a comparable dice score of 0.772+/-0.030 to a standalone segmentation model. The code is publicly available.
Segmentation with Super Images: A New 2D Perspective on 3D Medical Image Analysis
May 05, 2022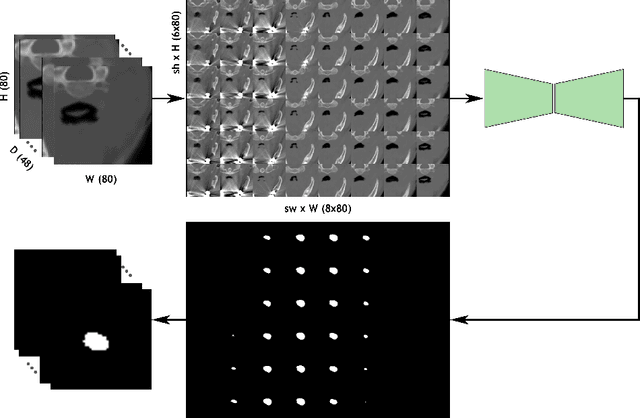
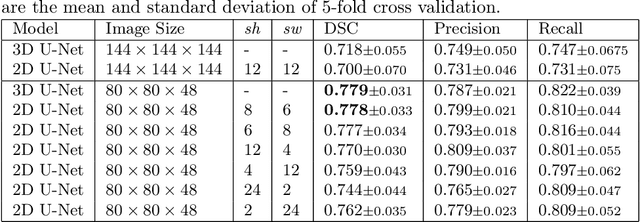

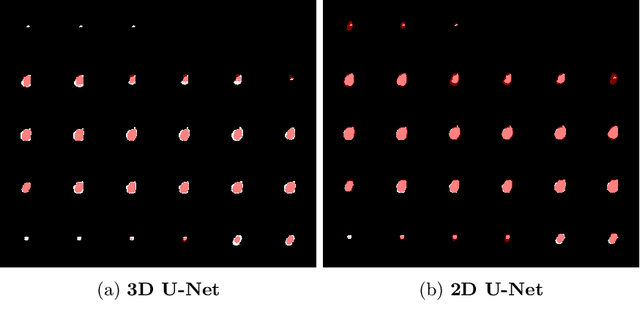
Abstract:Deep learning is showing an increasing number of audience in medical imaging research. In the segmentation task of medical images, we oftentimes rely on volumetric data, and thus require the use of 3D architectures which are praised for their ability to capture more features from the depth dimension. Yet, these architectures are generally more ineffective in time and compute compared to their 2D counterpart on account of 3D convolutions, max pooling, up-convolutions, and other operations used in these networks. Moreover, there are limited to no 3D pretrained model weights, and pretraining is generally challenging. To alleviate these issues, we propose to cast volumetric data to 2D super images and use 2D networks for the segmentation task. The method processes the 3D image by stitching slices side-by-side to generate a super resolution image. While the depth information is lost, we expect that deep neural networks can still capture and learn these features. Our goal in this work is to introduce a new perspective when dealing with volumetric data, and test our hypothesis using vanilla networks. We hope that this approach, while achieving close enough results to 3D networks using only 2D counterparts, can attract more related research in the future, especially in medical image analysis since volumetric data is comparably limited.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge