Hussain Alasmawi
FETAL-GAUGE: A Benchmark for Assessing Vision-Language Models in Fetal Ultrasound
Dec 25, 2025Abstract:The growing demand for prenatal ultrasound imaging has intensified a global shortage of trained sonographers, creating barriers to essential fetal health monitoring. Deep learning has the potential to enhance sonographers' efficiency and support the training of new practitioners. Vision-Language Models (VLMs) are particularly promising for ultrasound interpretation, as they can jointly process images and text to perform multiple clinical tasks within a single framework. However, despite the expansion of VLMs, no standardized benchmark exists to evaluate their performance in fetal ultrasound imaging. This gap is primarily due to the modality's challenging nature, operator dependency, and the limited public availability of datasets. To address this gap, we present Fetal-Gauge, the first and largest visual question answering benchmark specifically designed to evaluate VLMs across various fetal ultrasound tasks. Our benchmark comprises over 42,000 images and 93,000 question-answer pairs, spanning anatomical plane identification, visual grounding of anatomical structures, fetal orientation assessment, clinical view conformity, and clinical diagnosis. We systematically evaluate several state-of-the-art VLMs, including general-purpose and medical-specific models, and reveal a substantial performance gap: the best-performing model achieves only 55\% accuracy, far below clinical requirements. Our analysis identifies critical limitations of current VLMs in fetal ultrasound interpretation, highlighting the urgent need for domain-adapted architectures and specialized training approaches. Fetal-Gauge establishes a rigorous foundation for advancing multimodal deep learning in prenatal care and provides a pathway toward addressing global healthcare accessibility challenges. Our benchmark will be publicly available once the paper gets accepted.
crossMoDA Challenge: Evolution of Cross-Modality Domain Adaptation Techniques for Vestibular Schwannoma and Cochlea Segmentation from 2021 to 2023
Jun 13, 2025Abstract:The cross-Modality Domain Adaptation (crossMoDA) challenge series, initiated in 2021 in conjunction with the International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI), focuses on unsupervised cross-modality segmentation, learning from contrast-enhanced T1 (ceT1) and transferring to T2 MRI. The task is an extreme example of domain shift chosen to serve as a meaningful and illustrative benchmark. From a clinical application perspective, it aims to automate Vestibular Schwannoma (VS) and cochlea segmentation on T2 scans for more cost-effective VS management. Over time, the challenge objectives have evolved to enhance its clinical relevance. The challenge evolved from using single-institutional data and basic segmentation in 2021 to incorporating multi-institutional data and Koos grading in 2022, and by 2023, it included heterogeneous routine data and sub-segmentation of intra- and extra-meatal tumour components. In this work, we report the findings of the 2022 and 2023 editions and perform a retrospective analysis of the challenge progression over the years. The observations from the successive challenge contributions indicate that the number of outliers decreases with an expanding dataset. This is notable since the diversity of scanning protocols of the datasets concurrently increased. The winning approach of the 2023 edition reduced the number of outliers on the 2021 and 2022 testing data, demonstrating how increased data heterogeneity can enhance segmentation performance even on homogeneous data. However, the cochlea Dice score declined in 2023, likely due to the added complexity from tumour sub-annotations affecting overall segmentation performance. While progress is still needed for clinically acceptable VS segmentation, the plateauing performance suggests that a more challenging cross-modal task may better serve future benchmarking.
FetalCLIP: A Visual-Language Foundation Model for Fetal Ultrasound Image Analysis
Feb 20, 2025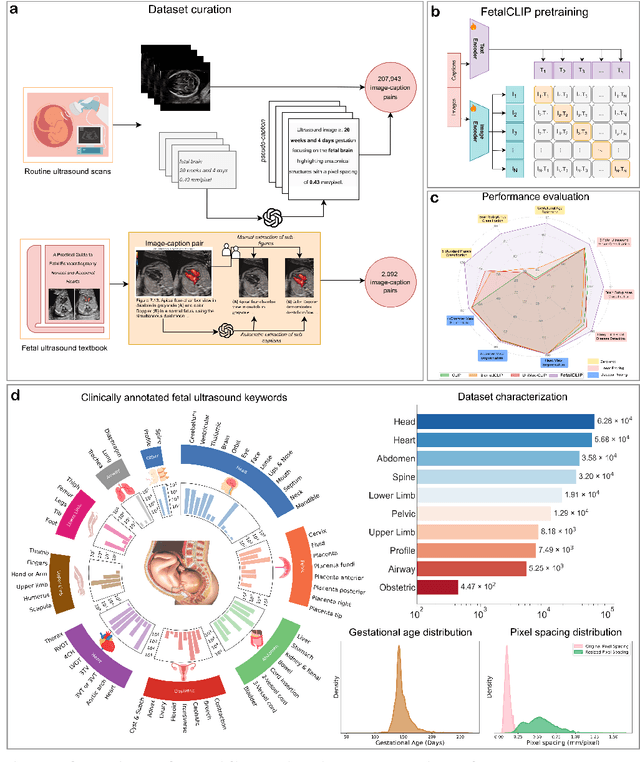
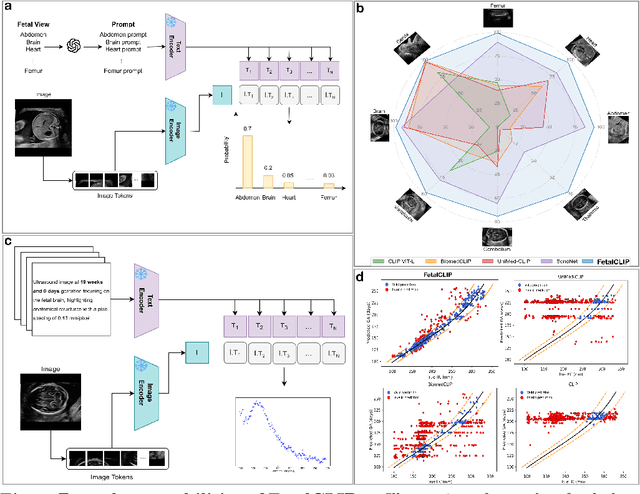
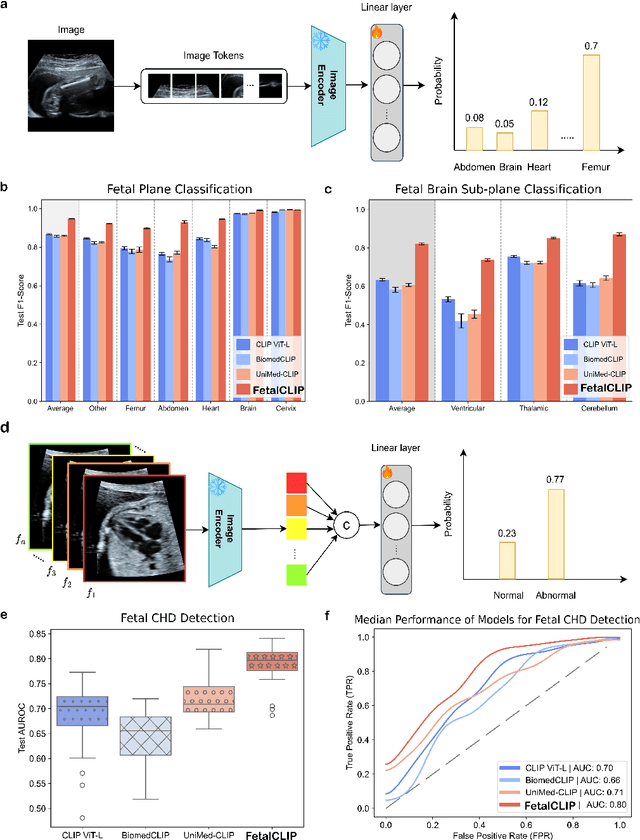
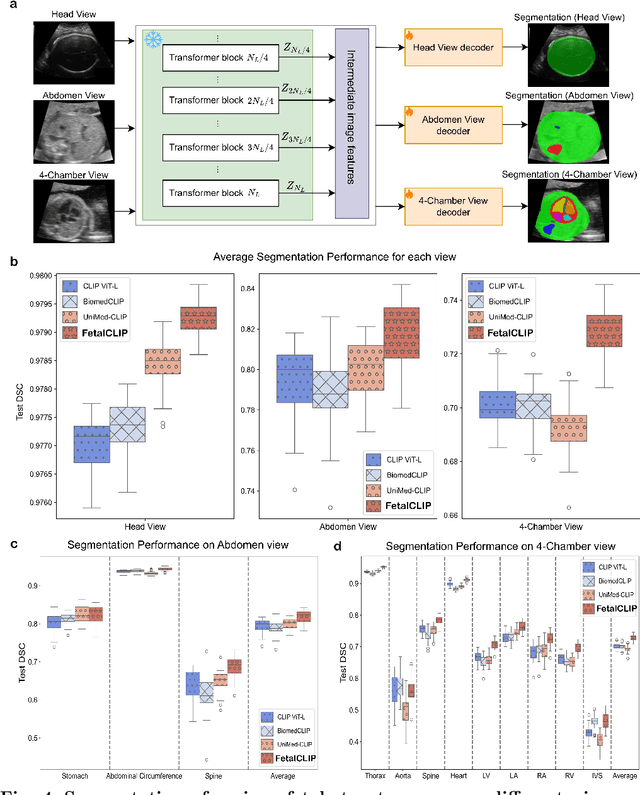
Abstract:Foundation models are becoming increasingly effective in the medical domain, offering pre-trained models on large datasets that can be readily adapted for downstream tasks. Despite progress, fetal ultrasound images remain a challenging domain for foundation models due to their inherent complexity, often requiring substantial additional training and facing limitations due to the scarcity of paired multimodal data. To overcome these challenges, here we introduce FetalCLIP, a vision-language foundation model capable of generating universal representation of fetal ultrasound images. FetalCLIP was pre-trained using a multimodal learning approach on a diverse dataset of 210,035 fetal ultrasound images paired with text. This represents the largest paired dataset of its kind used for foundation model development to date. This unique training approach allows FetalCLIP to effectively learn the intricate anatomical features present in fetal ultrasound images, resulting in robust representations that can be used for a variety of downstream applications. In extensive benchmarking across a range of key fetal ultrasound applications, including classification, gestational age estimation, congenital heart defect (CHD) detection, and fetal structure segmentation, FetalCLIP outperformed all baselines while demonstrating remarkable generalizability and strong performance even with limited labeled data. We plan to release the FetalCLIP model publicly for the benefit of the broader scientific community.
Leveraging Self-Supervised Learning for Fetal Cardiac Planes Classification using Ultrasound Scan Videos
Jul 31, 2024Abstract:Self-supervised learning (SSL) methods are popular since they can address situations with limited annotated data by directly utilising the underlying data distribution. However, the adoption of such methods is not explored enough in ultrasound (US) imaging, especially for fetal assessment. We investigate the potential of dual-encoder SSL in utilizing unlabelled US video data to improve the performance of challenging downstream Standard Fetal Cardiac Planes (SFCP) classification using limited labelled 2D US images. We study 7 SSL approaches based on reconstruction, contrastive loss, distillation, and information theory and evaluate them extensively on a large private US dataset. Our observations and findings are consolidated from more than 500 downstream training experiments under different settings. Our primary observation shows that for SSL training, the variance of the dataset is more crucial than its size because it allows the model to learn generalisable representations, which improve the performance of downstream tasks. Overall, the BarlowTwins method shows robust performance, irrespective of the training settings and data variations, when used as an initialisation for downstream tasks. Notably, full fine-tuning with 1% of labelled data outperforms ImageNet initialisation by 12% in F1-score and outperforms other SSL initialisations by at least 4% in F1-score, thus making it a promising candidate for transfer learning from US video to image data.
SurvRNC: Learning Ordered Representations for Survival Prediction using Rank-N-Contrast
Mar 15, 2024



Abstract:Predicting the likelihood of survival is of paramount importance for individuals diagnosed with cancer as it provides invaluable information regarding prognosis at an early stage. This knowledge enables the formulation of effective treatment plans that lead to improved patient outcomes. In the past few years, deep learning models have provided a feasible solution for assessing medical images, electronic health records, and genomic data to estimate cancer risk scores. However, these models often fall short of their potential because they struggle to learn regression-aware feature representations. In this study, we propose Survival Rank-N Contrast (SurvRNC) method, which introduces a loss function as a regularizer to obtain an ordered representation based on the survival times. This function can handle censored data and can be incorporated into any survival model to ensure that the learned representation is ordinal. The model was extensively evaluated on a HEad \& NeCK TumOR (HECKTOR) segmentation and the outcome-prediction task dataset. We demonstrate that using the SurvRNC method for training can achieve higher performance on different deep survival models. Additionally, it outperforms state-of-the-art methods by 3.6% on the concordance index. The code is publicly available on https://github.com/numanai/SurvRNC
FUSC: Fetal Ultrasound Semantic Clustering of Second Trimester Scans Using Deep Self-supervised Learning
Oct 19, 2023Abstract:Ultrasound is the primary imaging modality in clinical practice during pregnancy. More than 140M fetuses are born yearly, resulting in numerous scans. The availability of a large volume of fetal ultrasound scans presents the opportunity to train robust machine learning models. However, the abundance of scans also has its challenges, as manual labeling of each image is needed for supervised methods. Labeling is typically labor-intensive and requires expertise to annotate the images accurately. This study presents an unsupervised approach for automatically clustering ultrasound images into a large range of fetal views, reducing or eliminating the need for manual labeling. Our Fetal Ultrasound Semantic Clustering (FUSC) method is developed using a large dataset of 88,063 images and further evaluated on an additional unseen dataset of 8,187 images achieving over 92% clustering purity. The result of our investigation hold the potential to significantly impact the field of fetal ultrasound imaging and pave the way for more advanced automated labeling solutions. Finally, we make the code and the experimental setup publicly available to help advance the field.
MGMT promoter methylation status prediction using MRI scans? An extensive experimental evaluation of deep learning models
Apr 03, 2023Abstract:The number of studies on deep learning for medical diagnosis is expanding, and these systems are often claimed to outperform clinicians. However, only a few systems have shown medical efficacy. From this perspective, we examine a wide range of deep learning algorithms for the assessment of glioblastoma - a common brain tumor in older adults that is lethal. Surgery, chemotherapy, and radiation are the standard treatments for glioblastoma patients. The methylation status of the MGMT promoter, a specific genetic sequence found in the tumor, affects chemotherapy's effectiveness. MGMT promoter methylation improves chemotherapy response and survival in several cancers. MGMT promoter methylation is determined by a tumor tissue biopsy, which is then genetically tested. This lengthy and invasive procedure increases the risk of infection and other complications. Thus, researchers have used deep learning models to examine the tumor from brain MRI scans to determine the MGMT promoter's methylation state. We employ deep learning models and one of the largest public MRI datasets of 585 participants to predict the methylation status of the MGMT promoter in glioblastoma tumors using MRI scans. We test these models using Grad-CAM, occlusion sensitivity, feature visualizations, and training loss landscapes. Our results show no correlation between these two, indicating that external cohort data should be used to verify these models' performance to assure the accuracy and reliability of deep learning systems in cancer diagnosis.
Weakly Unsupervised Domain Adaptation for Vestibular Schwannoma Segmentation
Mar 13, 2023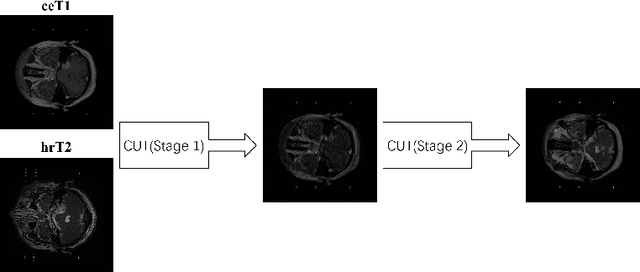



Abstract:Vestibular schwannoma (VS) is a non-cancerous tumor located next to the ear that can cause hearing loss. Most brain MRI images acquired from patients are contrast-enhanced T1 (ceT1), with a growing interest in high-resolution T2 images (hrT2) to replace ceT1, which involves the use of a contrast agent. As hrT2 images are currently scarce, it is less likely to train robust machine learning models to segment VS or other brain structures. In this work, we propose a weakly supervised machine learning approach that learns from only ceT1 scans and adapts to segment two structures from hrT2 scans: the VS and the cochlea from the crossMoDA dataset. Our model 1) generates fake hrT2 scans from ceT1 images and segmentation masks, 2) is trained using the fake hrT2 scans, 3) predicts the augmented real hrT2 scans, and 4) is retrained again using both the fake and real hrT2. The final result of this model has been computed on an unseen testing dataset provided by the 2022 crossMoDA challenge organizers. The mean dice score and average symmetric surface distance (ASSD) are 0.78 and 0.46, respectively. The predicted segmentation masks achieved a dice score of 0.83 and an ASSD of 0.56 on the VS, and a dice score of 0.74 and an ASSD of 0.35 on the cochleas.
Automatic Segmentation of Head and Neck Tumor: How Powerful Transformers Are?
Jan 17, 2022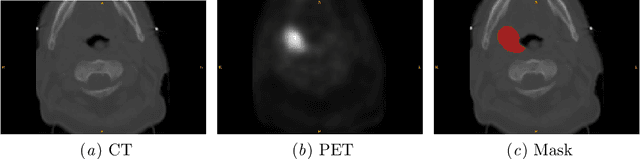

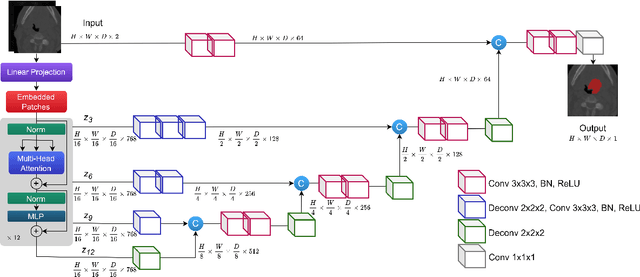
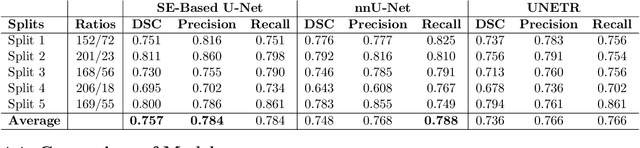
Abstract:Cancer is one of the leading causes of death worldwide, and head and neck (H&N) cancer is amongst the most prevalent types. Positron emission tomography and computed tomography are used to detect and segment the tumor region. Clinically, tumor segmentation is extensively time-consuming and prone to error. Machine learning, and deep learning in particular, can assist to automate this process, yielding results as accurate as the results of a clinician. In this research study, we develop a vision transformers-based method to automatically delineate H&N tumor, and compare its results to leading convolutional neural network (CNN)-based models. We use multi-modal data of CT and PET scans to do this task. We show that the selected transformer-based model can achieve results on a par with CNN-based ones. With cross validation, the model achieves a mean dice similarity coefficient of 0.736, mean precision of 0.766 and mean recall of 0.766. This is only 0.021 less than the 2020 competition winning model in terms of the DSC score. This indicates that the exploration of transformer-based models is a promising research area.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge