Leanne Bricker
FetalCLIP: A Visual-Language Foundation Model for Fetal Ultrasound Image Analysis
Feb 20, 2025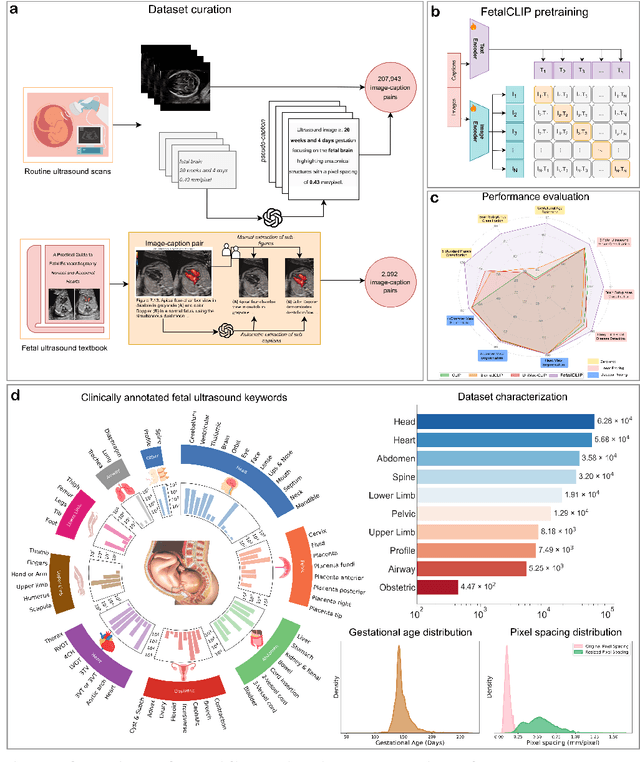
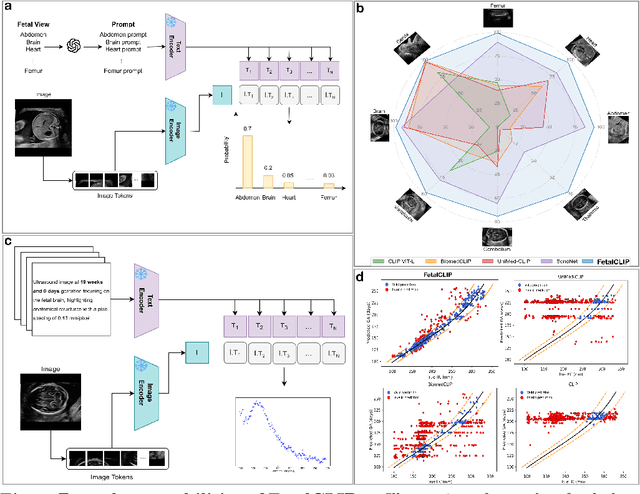
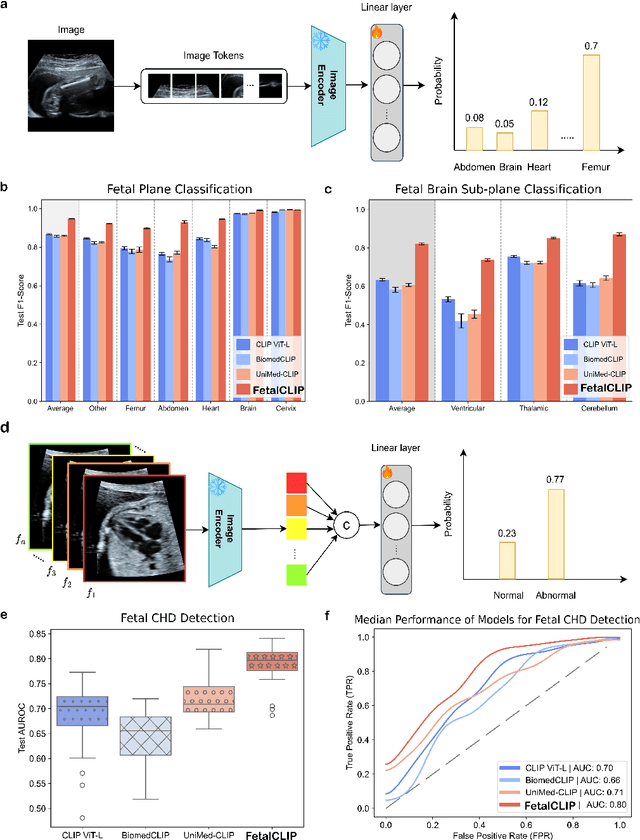
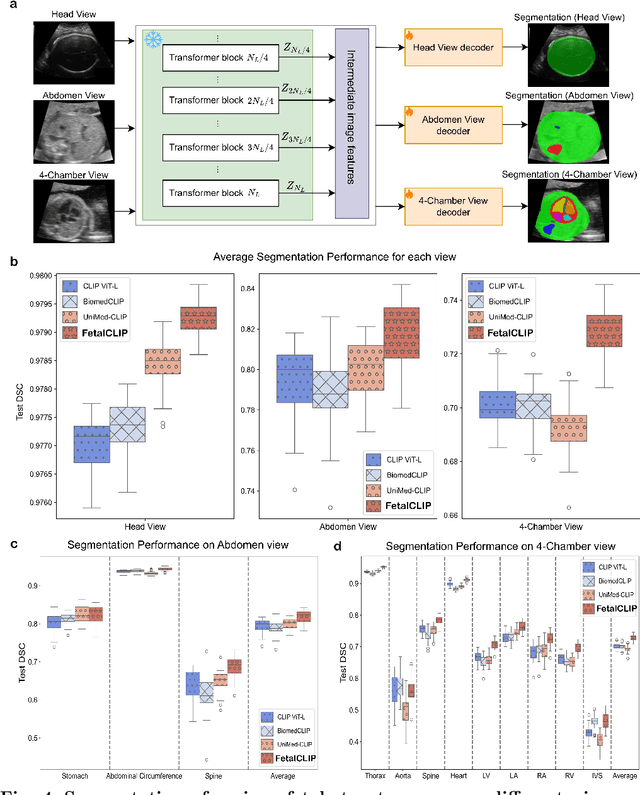
Abstract:Foundation models are becoming increasingly effective in the medical domain, offering pre-trained models on large datasets that can be readily adapted for downstream tasks. Despite progress, fetal ultrasound images remain a challenging domain for foundation models due to their inherent complexity, often requiring substantial additional training and facing limitations due to the scarcity of paired multimodal data. To overcome these challenges, here we introduce FetalCLIP, a vision-language foundation model capable of generating universal representation of fetal ultrasound images. FetalCLIP was pre-trained using a multimodal learning approach on a diverse dataset of 210,035 fetal ultrasound images paired with text. This represents the largest paired dataset of its kind used for foundation model development to date. This unique training approach allows FetalCLIP to effectively learn the intricate anatomical features present in fetal ultrasound images, resulting in robust representations that can be used for a variety of downstream applications. In extensive benchmarking across a range of key fetal ultrasound applications, including classification, gestational age estimation, congenital heart defect (CHD) detection, and fetal structure segmentation, FetalCLIP outperformed all baselines while demonstrating remarkable generalizability and strong performance even with limited labeled data. We plan to release the FetalCLIP model publicly for the benefit of the broader scientific community.
Leveraging Self-Supervised Learning for Fetal Cardiac Planes Classification using Ultrasound Scan Videos
Jul 31, 2024Abstract:Self-supervised learning (SSL) methods are popular since they can address situations with limited annotated data by directly utilising the underlying data distribution. However, the adoption of such methods is not explored enough in ultrasound (US) imaging, especially for fetal assessment. We investigate the potential of dual-encoder SSL in utilizing unlabelled US video data to improve the performance of challenging downstream Standard Fetal Cardiac Planes (SFCP) classification using limited labelled 2D US images. We study 7 SSL approaches based on reconstruction, contrastive loss, distillation, and information theory and evaluate them extensively on a large private US dataset. Our observations and findings are consolidated from more than 500 downstream training experiments under different settings. Our primary observation shows that for SSL training, the variance of the dataset is more crucial than its size because it allows the model to learn generalisable representations, which improve the performance of downstream tasks. Overall, the BarlowTwins method shows robust performance, irrespective of the training settings and data variations, when used as an initialisation for downstream tasks. Notably, full fine-tuning with 1% of labelled data outperforms ImageNet initialisation by 12% in F1-score and outperforms other SSL initialisations by at least 4% in F1-score, thus making it a promising candidate for transfer learning from US video to image data.
Multi-Task Learning Approach for Unified Biometric Estimation from Fetal Ultrasound Anomaly Scans
Nov 16, 2023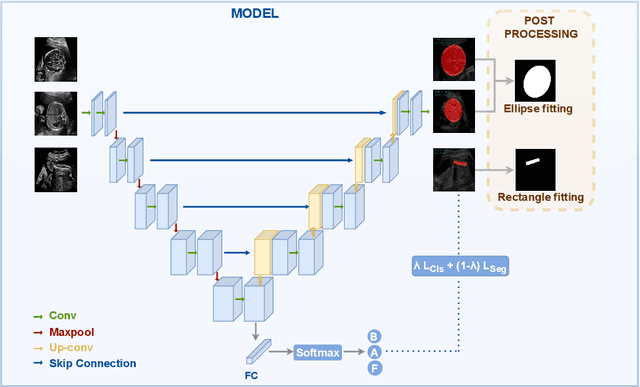
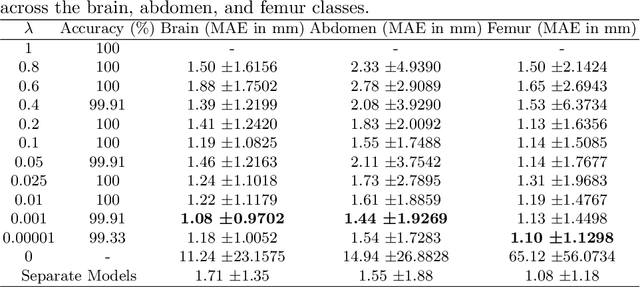

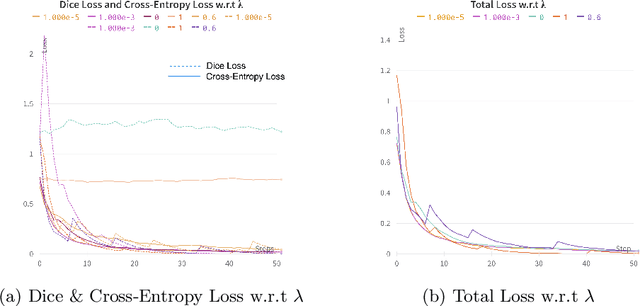
Abstract:Precise estimation of fetal biometry parameters from ultrasound images is vital for evaluating fetal growth, monitoring health, and identifying potential complications reliably. However, the automated computerized segmentation of the fetal head, abdomen, and femur from ultrasound images, along with the subsequent measurement of fetal biometrics, remains challenging. In this work, we propose a multi-task learning approach to classify the region into head, abdomen and femur as well as estimate the associated parameters. We were able to achieve a mean absolute error (MAE) of 1.08 mm on head circumference, 1.44 mm on abdomen circumference and 1.10 mm on femur length with a classification accuracy of 99.91\% on a dataset of fetal Ultrasound images. To achieve this, we leverage a weighted joint classification and segmentation loss function to train a U-Net architecture with an added classification head. The code can be accessed through \href{https://github.com/BioMedIA-MBZUAI/Multi-Task-Learning-Approach-for-Unified-Biometric-Estimation-from-Fetal-Ultrasound-Anomaly-Scans.git}{\texttt{Github}
FUSC: Fetal Ultrasound Semantic Clustering of Second Trimester Scans Using Deep Self-supervised Learning
Oct 19, 2023Abstract:Ultrasound is the primary imaging modality in clinical practice during pregnancy. More than 140M fetuses are born yearly, resulting in numerous scans. The availability of a large volume of fetal ultrasound scans presents the opportunity to train robust machine learning models. However, the abundance of scans also has its challenges, as manual labeling of each image is needed for supervised methods. Labeling is typically labor-intensive and requires expertise to annotate the images accurately. This study presents an unsupervised approach for automatically clustering ultrasound images into a large range of fetal views, reducing or eliminating the need for manual labeling. Our Fetal Ultrasound Semantic Clustering (FUSC) method is developed using a large dataset of 88,063 images and further evaluated on an additional unseen dataset of 8,187 images achieving over 92% clustering purity. The result of our investigation hold the potential to significantly impact the field of fetal ultrasound imaging and pave the way for more advanced automated labeling solutions. Finally, we make the code and the experimental setup publicly available to help advance the field.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge