Werner Gerhard Diehl
Leveraging Self-Supervised Learning for Fetal Cardiac Planes Classification using Ultrasound Scan Videos
Jul 31, 2024Abstract:Self-supervised learning (SSL) methods are popular since they can address situations with limited annotated data by directly utilising the underlying data distribution. However, the adoption of such methods is not explored enough in ultrasound (US) imaging, especially for fetal assessment. We investigate the potential of dual-encoder SSL in utilizing unlabelled US video data to improve the performance of challenging downstream Standard Fetal Cardiac Planes (SFCP) classification using limited labelled 2D US images. We study 7 SSL approaches based on reconstruction, contrastive loss, distillation, and information theory and evaluate them extensively on a large private US dataset. Our observations and findings are consolidated from more than 500 downstream training experiments under different settings. Our primary observation shows that for SSL training, the variance of the dataset is more crucial than its size because it allows the model to learn generalisable representations, which improve the performance of downstream tasks. Overall, the BarlowTwins method shows robust performance, irrespective of the training settings and data variations, when used as an initialisation for downstream tasks. Notably, full fine-tuning with 1% of labelled data outperforms ImageNet initialisation by 12% in F1-score and outperforms other SSL initialisations by at least 4% in F1-score, thus making it a promising candidate for transfer learning from US video to image data.
Multi-Task Learning Approach for Unified Biometric Estimation from Fetal Ultrasound Anomaly Scans
Nov 16, 2023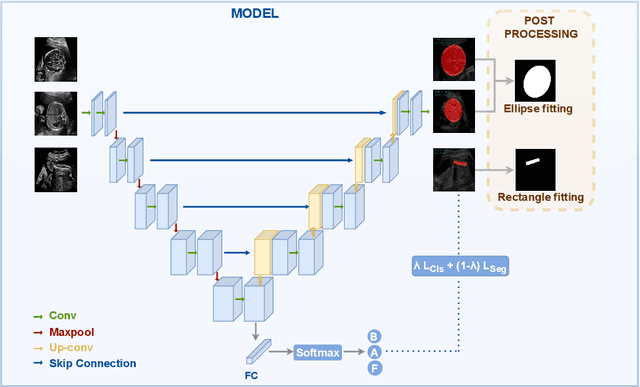
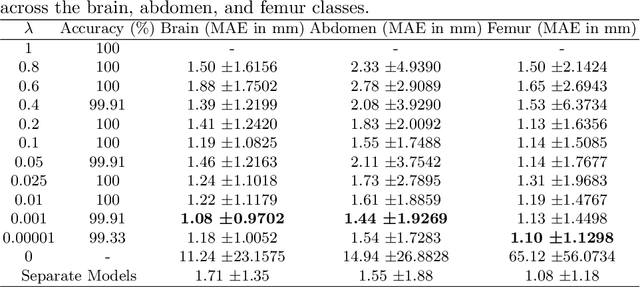

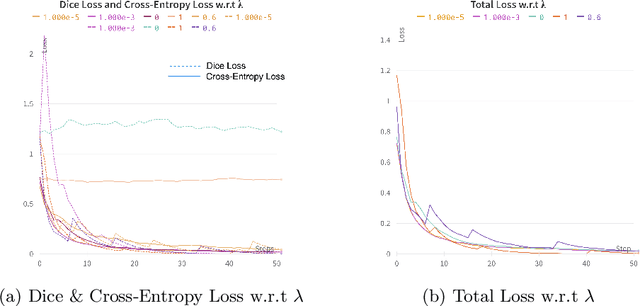
Abstract:Precise estimation of fetal biometry parameters from ultrasound images is vital for evaluating fetal growth, monitoring health, and identifying potential complications reliably. However, the automated computerized segmentation of the fetal head, abdomen, and femur from ultrasound images, along with the subsequent measurement of fetal biometrics, remains challenging. In this work, we propose a multi-task learning approach to classify the region into head, abdomen and femur as well as estimate the associated parameters. We were able to achieve a mean absolute error (MAE) of 1.08 mm on head circumference, 1.44 mm on abdomen circumference and 1.10 mm on femur length with a classification accuracy of 99.91\% on a dataset of fetal Ultrasound images. To achieve this, we leverage a weighted joint classification and segmentation loss function to train a U-Net architecture with an added classification head. The code can be accessed through \href{https://github.com/BioMedIA-MBZUAI/Multi-Task-Learning-Approach-for-Unified-Biometric-Estimation-from-Fetal-Ultrasound-Anomaly-Scans.git}{\texttt{Github}
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge