Mohammed Talha Alam
Quran-MD: A Fine-Grained Multilingual Multimodal Dataset of the Quran
Jan 25, 2026Abstract:We present Quran MD, a comprehensive multimodal dataset of the Quran that integrates textual, linguistic, and audio dimensions at the verse and word levels. For each verse (ayah), the dataset provides its original Arabic text, English translation, and phonetic transliteration. To capture the rich oral tradition of Quranic recitation, we include verse-level audio from 32 distinct reciters, reflecting diverse recitation styles and dialectical nuances. At the word level, each token is paired with its corresponding Arabic script, English translation, transliteration, and an aligned audio recording, allowing fine-grained analysis of pronunciation, phonology, and semantic context. This dataset supports various applications, including natural language processing, speech recognition, text-to-speech synthesis, linguistic analysis, and digital Islamic studies. Bridging text and audio modalities across multiple reciters, this dataset provides a unique resource to advance computational approaches to Quranic recitation and study. Beyond enabling tasks such as ASR, tajweed detection, and Quranic TTS, it lays the foundation for multimodal embeddings, semantic retrieval, style transfer, and personalized tutoring systems that can support both research and community applications. The dataset is available at https://huggingface.co/datasets/Buraaq/quran-audio-text-dataset
AdaptPrompt: Parameter-Efficient Adaptation of VLMs for Generalizable Deepfake Detection
Dec 19, 2025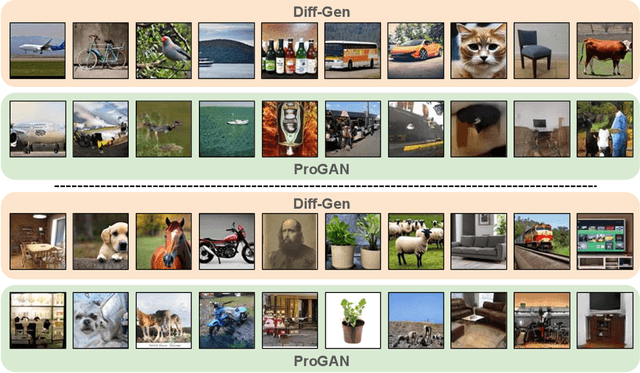
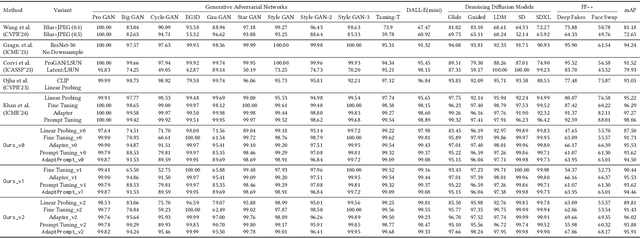
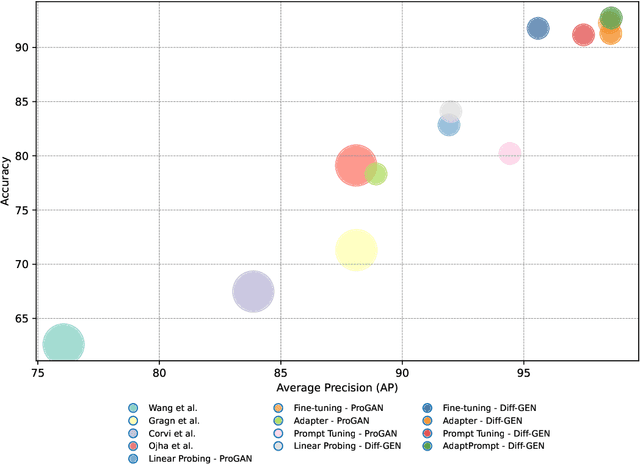
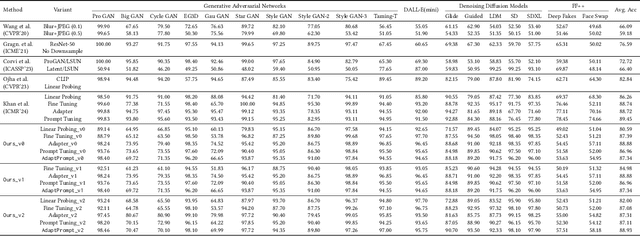
Abstract:Recent advances in image generation have led to the widespread availability of highly realistic synthetic media, increasing the difficulty of reliable deepfake detection. A key challenge is generalization, as detectors trained on a narrow class of generators often fail when confronted with unseen models. In this work, we address the pressing need for generalizable detection by leveraging large vision-language models, specifically CLIP, to identify synthetic content across diverse generative techniques. First, we introduce Diff-Gen, a large-scale benchmark dataset comprising 100k diffusion-generated fakes that capture broad spectral artifacts unlike traditional GAN datasets. Models trained on Diff-Gen demonstrate stronger cross-domain generalization, particularly on previously unseen image generators. Second, we propose AdaptPrompt, a parameter-efficient transfer learning framework that jointly learns task-specific textual prompts and visual adapters while keeping the CLIP backbone frozen. We further show via layer ablation that pruning the final transformer block of the vision encoder enhances the retention of high-frequency generative artifacts, significantly boosting detection accuracy. Our evaluation spans 25 challenging test sets, covering synthetic content generated by GANs, diffusion models, and commercial tools, establishing a new state-of-the-art in both standard and cross-domain scenarios. We further demonstrate the framework's versatility through few-shot generalization (using as few as 320 images) and source attribution, enabling the precise identification of generator architectures in closed-set settings.
Robust and Calibrated Detection of Authentic Multimedia Content
Dec 17, 2025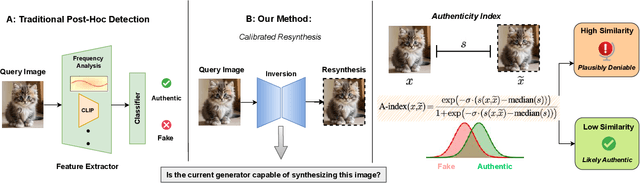
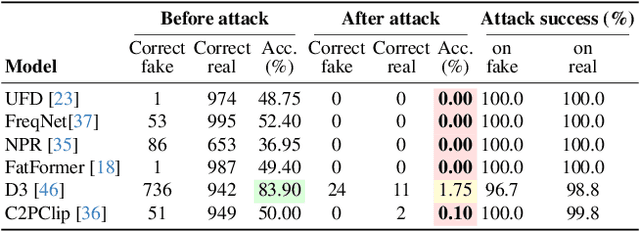
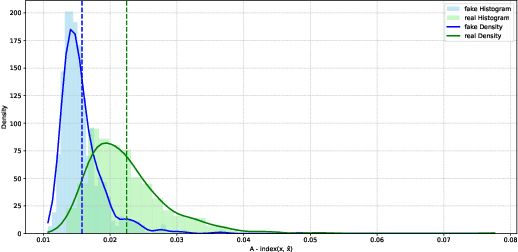
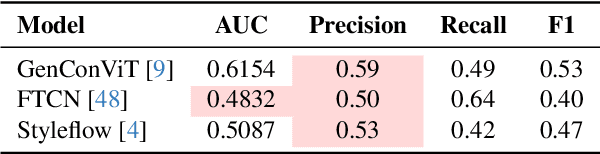
Abstract:Generative models can synthesize highly realistic content, so-called deepfakes, that are already being misused at scale to undermine digital media authenticity. Current deepfake detection methods are unreliable for two reasons: (i) distinguishing inauthentic content post-hoc is often impossible (e.g., with memorized samples), leading to an unbounded false positive rate (FPR); and (ii) detection lacks robustness, as adversaries can adapt to known detectors with near-perfect accuracy using minimal computational resources. To address these limitations, we propose a resynthesis framework to determine if a sample is authentic or if its authenticity can be plausibly denied. We make two key contributions focusing on the high-precision, low-recall setting against efficient (i.e., compute-restricted) adversaries. First, we demonstrate that our calibrated resynthesis method is the most reliable approach for verifying authentic samples while maintaining controllable, low FPRs. Second, we show that our method achieves adversarial robustness against efficient adversaries, whereas prior methods are easily evaded under identical compute budgets. Our approach supports multiple modalities and leverages state-of-the-art inversion techniques.
FaceAnonyMixer: Cancelable Faces via Identity Consistent Latent Space Mixing
Aug 07, 2025Abstract:Advancements in face recognition (FR) technologies have amplified privacy concerns, necessitating methods that protect identity while maintaining recognition utility. Existing face anonymization methods typically focus on obscuring identity but fail to meet the requirements of biometric template protection, including revocability, unlinkability, and irreversibility. We propose FaceAnonyMixer, a cancelable face generation framework that leverages the latent space of a pre-trained generative model to synthesize privacy-preserving face images. The core idea of FaceAnonyMixer is to irreversibly mix the latent code of a real face image with a synthetic code derived from a revocable key. The mixed latent code is further refined through a carefully designed multi-objective loss to satisfy all cancelable biometric requirements. FaceAnonyMixer is capable of generating high-quality cancelable faces that can be directly matched using existing FR systems without requiring any modifications. Extensive experiments on benchmark datasets demonstrate that FaceAnonyMixer delivers superior recognition accuracy while providing significantly stronger privacy protection, achieving over an 11% gain on commercial API compared to recent cancelable biometric methods. Code is available at: https://github.com/talha-alam/faceanonymixer.
Introducing SDICE: An Index for Assessing Diversity of Synthetic Medical Datasets
Sep 28, 2024



Abstract:Advancements in generative modeling are pushing the state-of-the-art in synthetic medical image generation. These synthetic images can serve as an effective data augmentation method to aid the development of more accurate machine learning models for medical image analysis. While the fidelity of these synthetic images has progressively increased, the diversity of these images is an understudied phenomenon. In this work, we propose the SDICE index, which is based on the characterization of similarity distributions induced by a contrastive encoder. Given a synthetic dataset and a reference dataset of real images, the SDICE index measures the distance between the similarity score distributions of original and synthetic images, where the similarity scores are estimated using a pre-trained contrastive encoder. This distance is then normalized using an exponential function to provide a consistent metric that can be easily compared across domains. Experiments conducted on the MIMIC-chest X-ray and ImageNet datasets demonstrate the effectiveness of SDICE index in assessing synthetic medical dataset diversity.
CosmoCLIP: Generalizing Large Vision-Language Models for Astronomical Imaging
Jul 10, 2024



Abstract:Existing vision-text contrastive learning models enhance representation transferability and support zero-shot prediction by matching paired image and caption embeddings while pushing unrelated pairs apart. However, astronomical image-label datasets are significantly smaller compared to general image and label datasets available from the internet. We introduce CosmoCLIP, an astronomical image-text contrastive learning framework precisely fine-tuned on the pre-trained CLIP model using SpaceNet and BLIP-based captions. SpaceNet, attained via FLARE, constitutes ~13k optimally distributed images, while BLIP acts as a rich knowledge extractor. The rich semantics derived from this SpaceNet and BLIP descriptions, when learned contrastively, enable CosmoCLIP to achieve superior generalization across various in-domain and out-of-domain tasks. Our results demonstrate that CosmoCLIP is a straightforward yet powerful framework, significantly outperforming CLIP in zero-shot classification and image-text retrieval tasks.
AstroSpy: On detecting Fake Images in Astronomy via Joint Image-Spectral Representations
Jul 09, 2024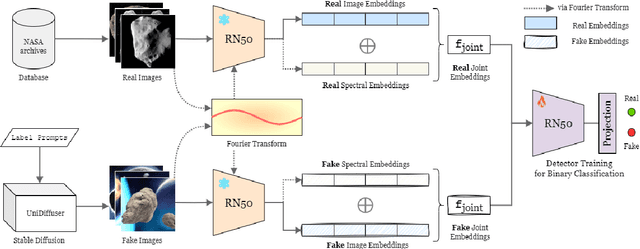
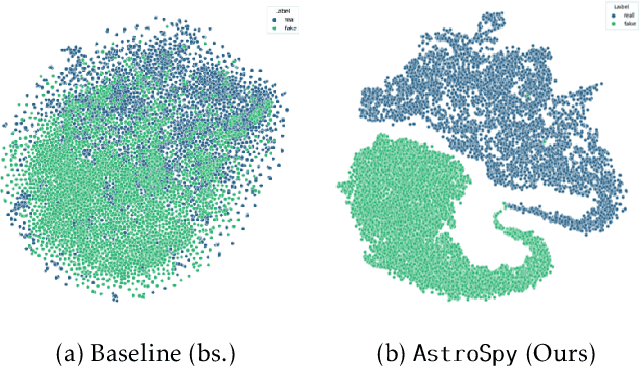
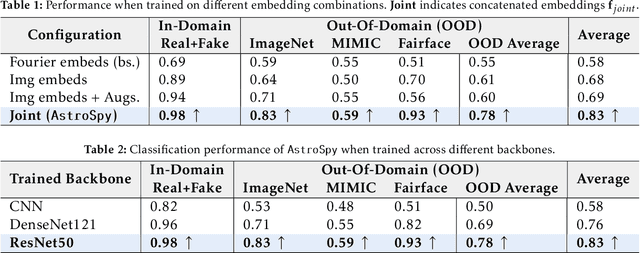
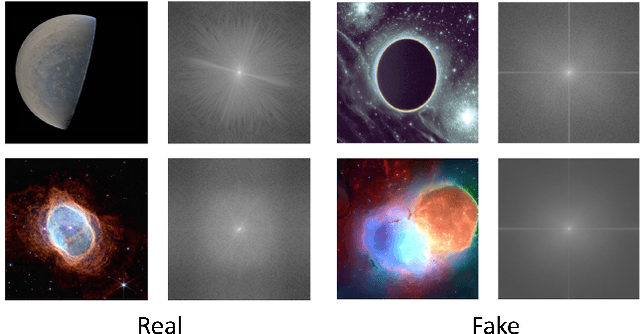
Abstract:The prevalence of AI-generated imagery has raised concerns about the authenticity of astronomical images, especially with advanced text-to-image models like Stable Diffusion producing highly realistic synthetic samples. Existing detection methods, primarily based on convolutional neural networks (CNNs) or spectral analysis, have limitations when used independently. We present AstroSpy, a hybrid model that integrates both spectral and image features to distinguish real from synthetic astronomical images. Trained on a unique dataset of real NASA images and AI-generated fakes (approximately 18k samples), AstroSpy utilizes a dual-pathway architecture to fuse spatial and spectral information. This approach enables AstroSpy to achieve superior performance in identifying authentic astronomical images. Extensive evaluations demonstrate AstroSpy's effectiveness and robustness, significantly outperforming baseline models in both in-domain and cross-domain tasks, highlighting its potential to combat misinformation in astronomy.
FLARE up your data: Diffusion-based Augmentation Method in Astronomical Imaging
May 22, 2024Abstract:The intersection of Astronomy and AI encounters significant challenges related to issues such as noisy backgrounds, lower resolution (LR), and the intricate process of filtering and archiving images from advanced telescopes like the James Webb. Given the dispersion of raw images in feature space, we have proposed a \textit{two-stage augmentation framework} entitled as \textbf{FLARE} based on \underline{f}eature \underline{l}earning and \underline{a}ugmented \underline{r}esolution \underline{e}nhancement. We first apply lower (LR) to higher resolution (HR) conversion followed by standard augmentations. Secondly, we integrate a diffusion approach to synthetically generate samples using class-concatenated prompts. By merging these two stages using weighted percentiles, we realign the feature space distribution, enabling a classification model to establish a distinct decision boundary and achieve superior generalization on various in-domain and out-of-domain tasks. We conducted experiments on several downstream cosmos datasets and on our optimally distributed \textbf{SpaceNet} dataset across 8-class fine-grained and 4-class macro classification tasks. FLARE attains the highest performance gain of 20.78\% for fine-grained tasks compared to similar baselines, while across different classification models, FLARE shows a consistent increment of an average of +15\%. This outcome underscores the effectiveness of the FLARE method in enhancing the precision of image classification, ultimately bolstering the reliability of astronomical research outcomes. % Our code and SpaceNet dataset will be released to the public soon. Our code and SpaceNet dataset is available at \href{https://github.com/Razaimam45/PlanetX_Dxb}{\textit{https://github.com/Razaimam45/PlanetX\_Dxb}}.
Multi-Task Learning Approach for Unified Biometric Estimation from Fetal Ultrasound Anomaly Scans
Nov 16, 2023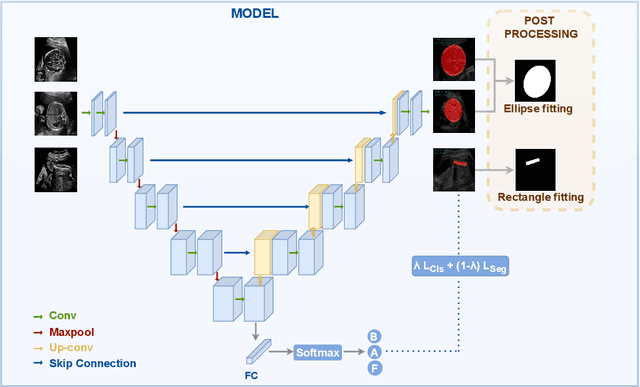
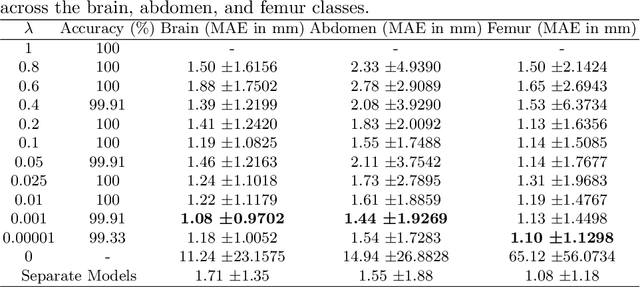

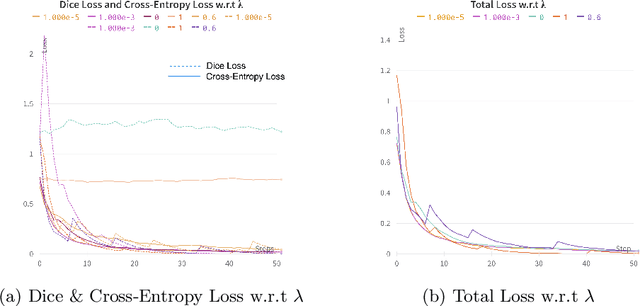
Abstract:Precise estimation of fetal biometry parameters from ultrasound images is vital for evaluating fetal growth, monitoring health, and identifying potential complications reliably. However, the automated computerized segmentation of the fetal head, abdomen, and femur from ultrasound images, along with the subsequent measurement of fetal biometrics, remains challenging. In this work, we propose a multi-task learning approach to classify the region into head, abdomen and femur as well as estimate the associated parameters. We were able to achieve a mean absolute error (MAE) of 1.08 mm on head circumference, 1.44 mm on abdomen circumference and 1.10 mm on femur length with a classification accuracy of 99.91\% on a dataset of fetal Ultrasound images. To achieve this, we leverage a weighted joint classification and segmentation loss function to train a U-Net architecture with an added classification head. The code can be accessed through \href{https://github.com/BioMedIA-MBZUAI/Multi-Task-Learning-Approach-for-Unified-Biometric-Estimation-from-Fetal-Ultrasound-Anomaly-Scans.git}{\texttt{Github}
Optimizing Brain Tumor Classification: A Comprehensive Study on Transfer Learning and Imbalance Handling in Deep Learning Models
Aug 13, 2023Abstract:Deep learning has emerged as a prominent field in recent literature, showcasing the introduction of models that utilize transfer learning to achieve remarkable accuracies in the classification of brain tumor MRI images. However, the majority of these proposals primarily focus on balanced datasets, neglecting the inherent data imbalance present in real-world scenarios. Consequently, there is a pressing need for approaches that not only address the data imbalance but also prioritize precise classification of brain cancer. In this work, we present a novel deep learning-based approach, called Transfer Learning-CNN, for brain tumor classification using MRI data. The proposed model leverages the predictive capabilities of existing publicly available models by utilizing their pre-trained weights and transferring those weights to the CNN. By leveraging a publicly available Brain MRI dataset, the experiment evaluated various transfer learning models for classifying different tumor types, including meningioma, glioma, and pituitary tumors. We investigate the impact of different loss functions, including focal loss, and oversampling methods, such as SMOTE and ADASYN, in addressing the data imbalance issue. Notably, the proposed strategy, which combines VGG-16 and CNN, achieved an impressive accuracy rate of 96%, surpassing alternative approaches significantly.
* Our code is available at https://github.com/Razaimam45/AI701-Project-Transfer-Learning-approach-for-imbalance-classification-of-Brain-Tumor-MRI-
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge