Ibrahim Almakky
MAFM^3: Modular Adaptation of Foundation Models for Multi-Modal Medical AI
Nov 14, 2025Abstract:Foundational models are trained on extensive datasets to capture the general trends of a domain. However, in medical imaging, the scarcity of data makes pre-training for every domain, modality, or task challenging. Instead of building separate models, we propose MAFM^3 (Modular Adaptation of Foundation Models for Multi-Modal Medical AI), a framework that enables a single foundation model to expand into diverse domains, tasks, and modalities through lightweight modular components. These components serve as specialized skill sets that allow the system to flexibly activate the appropriate capability at the inference time, depending on the input type or clinical objective. Unlike conventional adaptation methods that treat each new task or modality in isolation, MAFM^3 provides a unified and expandable framework for efficient multitask and multimodality adaptation. Empirically, we validate our approach by adapting a chest CT foundation model initially trained for classification into prognosis and segmentation modules. Our results show improved performance on both tasks. Furthermore, by incorporating PET scans, MAFM^3 achieved an improvement in the Dice score 5% compared to the respective baselines. These findings establish that foundation models, when equipped with modular components, are not inherently constrained to their initial training scope but can evolve into multitask, multimodality systems for medical imaging. The code implementation of this work can be found at https://github.com/Areeb2735/CTscan_prognosis_VLM
SALT: Singular Value Adaptation with Low-Rank Transformation
Mar 20, 2025Abstract:The complex nature of medical image segmentation calls for models that are specifically designed to capture detailed, domain-specific features. Large foundation models offer considerable flexibility, yet the cost of fine-tuning these models remains a significant barrier. Parameter-Efficient Fine-Tuning (PEFT) methods, such as Low-Rank Adaptation (LoRA), efficiently update model weights with low-rank matrices but may suffer from underfitting when the chosen rank is insufficient to capture domain-specific nuances. Conversely, full-rank Singular Value Decomposition (SVD) based methods provide comprehensive updates by modifying all singular values, yet they often lack flexibility and exhibit variable performance across datasets. We propose SALT (Singular Value Adaptation with Low-Rank Transformation), a method that selectively adapts the most influential singular values using trainable scale and shift parameters while complementing this with a low-rank update for the remaining subspace. This hybrid approach harnesses the advantages of both LoRA and SVD, enabling effective adaptation without relying on increasing model size or depth. Evaluated on 5 challenging medical datasets, ranging from as few as 20 samples to 1000, SALT outperforms state-of-the-art PEFT (LoRA and SVD) by 2% to 5% in Dice with only 3.9% trainable parameters, demonstrating robust adaptation even in low-resource settings. The code for SALT is available at: https://github.com/BioMedIA-MBZUAI/SALT
In-Model Merging for Enhancing the Robustness of Medical Imaging Classification Models
Feb 27, 2025



Abstract:Model merging is an effective strategy to merge multiple models for enhancing model performances, and more efficient than ensemble learning as it will not introduce extra computation into inference. However, limited research explores if the merging process can occur within one model and enhance the model's robustness, which is particularly critical in the medical image domain. In the paper, we are the first to propose in-model merging (InMerge), a novel approach that enhances the model's robustness by selectively merging similar convolutional kernels in the deep layers of a single convolutional neural network (CNN) during the training process for classification. We also analytically reveal important characteristics that affect how in-model merging should be performed, serving as an insightful reference for the community. We demonstrate the feasibility and effectiveness of this technique for different CNN architectures on 4 prevalent datasets. The proposed InMerge-trained model surpasses the typically-trained model by a substantial margin. The code will be made public.
Rethinking Weight-Averaged Model-merging
Nov 14, 2024



Abstract:Weight-averaged model-merging has emerged as a powerful approach in deep learning, capable of enhancing model performance without fine-tuning or retraining. However, the underlying mechanisms that explain its effectiveness remain largely unexplored. In this paper, we investigate this technique from three novel perspectives to provide deeper insights into how and why weight-averaged model-merging works: (1) we examine the intrinsic patterns captured by the learning of the model weights, through the visualizations of their patterns on several datasets, showing that these weights often encode structured and interpretable patterns; (2) we investigate model ensemble merging strategies based on averaging on weights versus averaging on features, providing detailed analyses across diverse architectures and datasets; and (3) we explore the impact on model-merging prediction stability in terms of changing the parameter magnitude, revealing insights into the way of weight averaging works as regularization by showing the robustness across different parameter scales. Our findings shed light on the "black box" of weight-averaged model-merging, offering valuable insights and practical recommendations that advance the model-merging process.
Continual Learning in Medical Imaging from Theory to Practice: A Survey and Practical Analysis
May 22, 2024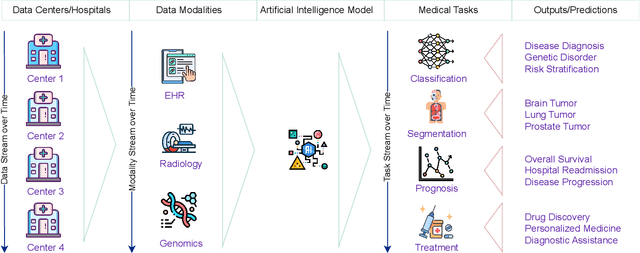
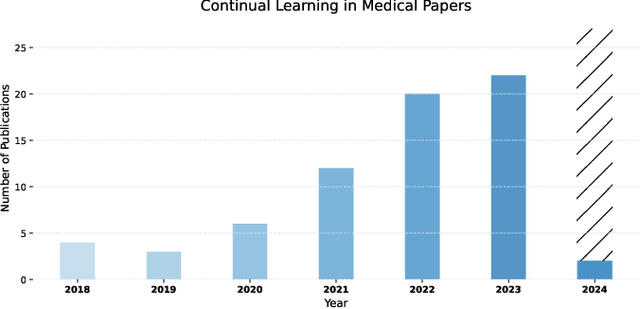

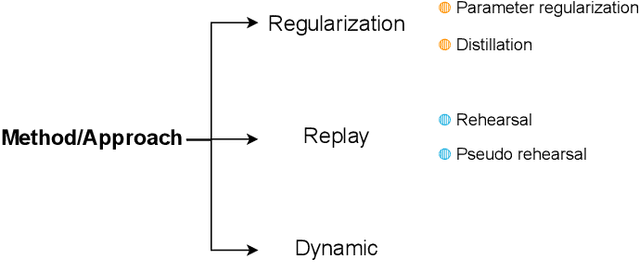
Abstract:Deep Learning has shown great success in reshaping medical imaging, yet it faces numerous challenges hindering widespread application. Issues like catastrophic forgetting and distribution shifts in the continuously evolving data stream increase the gap between research and applications. Continual Learning offers promise in addressing these hurdles by enabling the sequential acquisition of new knowledge without forgetting previous learnings in neural networks. In this survey, we comprehensively review the recent literature on continual learning in the medical domain, highlight recent trends, and point out the practical issues. Specifically, we survey the continual learning studies on classification, segmentation, detection, and other tasks in the medical domain. Furthermore, we develop a taxonomy for the reviewed studies, identify the challenges, and provide insights to overcome them. We also critically discuss the current state of continual learning in medical imaging, including identifying open problems and outlining promising future directions. We hope this survey will provide researchers with a useful overview of the developments in the field and will further increase interest in the community. To keep up with the fast-paced advancements in this field, we plan to routinely update the repository with the latest relevant papers at https://github.com/BioMedIA-MBZUAI/awesome-cl-in-medical .
DynaMMo: Dynamic Model Merging for Efficient Class Incremental Learning for Medical Images
Apr 22, 2024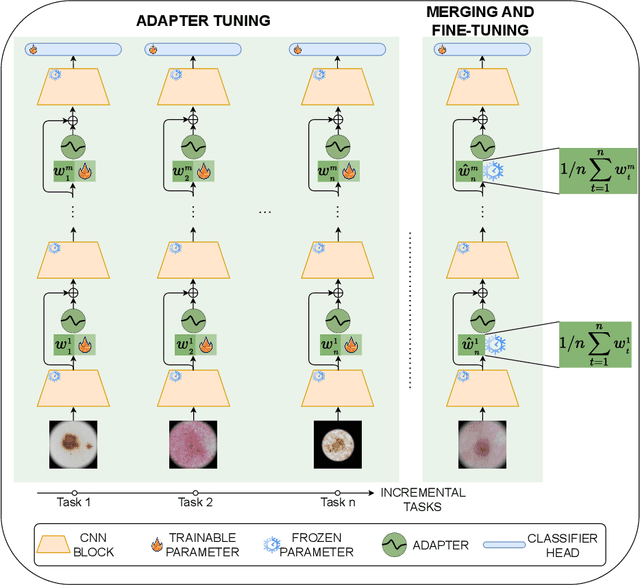
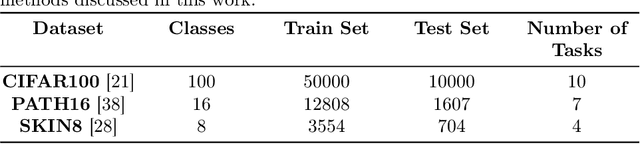
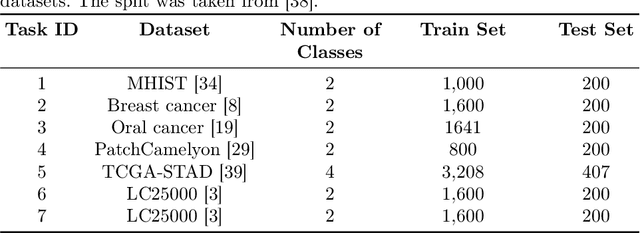
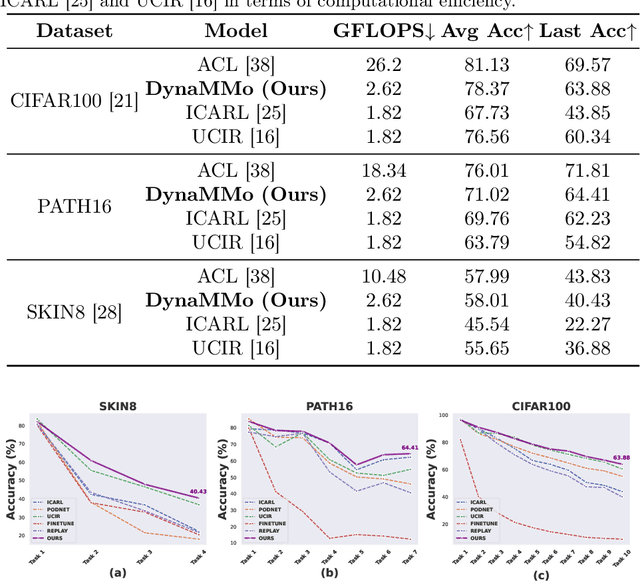
Abstract:Continual learning, the ability to acquire knowledge from new data while retaining previously learned information, is a fundamental challenge in machine learning. Various approaches, including memory replay, knowledge distillation, model regularization, and dynamic network expansion, have been proposed to address this issue. Thus far, dynamic network expansion methods have achieved state-of-the-art performance at the cost of incurring significant computational overhead. This is due to the need for additional model buffers, which makes it less feasible in resource-constrained settings, particularly in the medical domain. To overcome this challenge, we propose Dynamic Model Merging, DynaMMo, a method that merges multiple networks at different stages of model training to achieve better computational efficiency. Specifically, we employ lightweight learnable modules for each task and combine them into a unified model to minimize computational overhead. DynaMMo achieves this without compromising performance, offering a cost-effective solution for continual learning in medical applications. We evaluate DynaMMo on three publicly available datasets, demonstrating its effectiveness compared to existing approaches. DynaMMo offers around 10-fold reduction in GFLOPS with a small drop of 2.76 in average accuracy when compared to state-of-the-art dynamic-based approaches. The code implementation of this work will be available upon the acceptance of this work at https://github.com/BioMedIA-MBZUAI/DynaMMo.
FissionFusion: Fast Geometric Generation and Hierarchical Souping for Medical Image Analysis
Mar 20, 2024



Abstract:The scarcity of well-annotated medical datasets requires leveraging transfer learning from broader datasets like ImageNet or pre-trained models like CLIP. Model soups averages multiple fine-tuned models aiming to improve performance on In-Domain (ID) tasks and enhance robustness against Out-of-Distribution (OOD) datasets. However, applying these methods to the medical imaging domain faces challenges and results in suboptimal performance. This is primarily due to differences in error surface characteristics that stem from data complexities such as heterogeneity, domain shift, class imbalance, and distributional shifts between training and testing phases. To address this issue, we propose a hierarchical merging approach that involves local and global aggregation of models at various levels based on models' hyperparameter configurations. Furthermore, to alleviate the need for training a large number of models in the hyperparameter search, we introduce a computationally efficient method using a cyclical learning rate scheduler to produce multiple models for aggregation in the weight space. Our method demonstrates significant improvements over the model souping approach across multiple datasets (around 6% gain in HAM10000 and CheXpert datasets) while maintaining low computational costs for model generation and selection. Moreover, we achieve better results on OOD datasets than model soups. The code is available at https://github.com/BioMedIA-MBZUAI/FissionFusion.
TiBiX: Leveraging Temporal Information for Bidirectional X-ray and Report Generation
Mar 20, 2024



Abstract:With the emergence of vision language models in the medical imaging domain, numerous studies have focused on two dominant research activities: (1) report generation from Chest X-rays (CXR), and (2) synthetic scan generation from text or reports. Despite some research incorporating multi-view CXRs into the generative process, prior patient scans and reports have been generally disregarded. This can inadvertently lead to the leaving out of important medical information, thus affecting generation quality. To address this, we propose TiBiX: Leveraging Temporal information for Bidirectional X-ray and Report Generation. Considering previous scans, our approach facilitates bidirectional generation, primarily addressing two challenging problems: (1) generating the current image from the previous image and current report and (2) generating the current report based on both the previous and current images. Moreover, we extract and release a curated temporal benchmark dataset derived from the MIMIC-CXR dataset, which focuses on temporal data. Our comprehensive experiments and ablation studies explore the merits of incorporating prior CXRs and achieve state-of-the-art (SOTA) results on the report generation task. Furthermore, we attain on-par performance with SOTA image generation efforts, thus serving as a new baseline in longitudinal bidirectional CXR-to-report generation. The code is available at https://github.com/BioMedIA-MBZUAI/TiBiX.
MedMerge: Merging Models for Effective Transfer Learning to Medical Imaging Tasks
Mar 18, 2024



Abstract:Transfer learning has become a powerful tool to initialize deep learning models to achieve faster convergence and higher performance. This is especially useful in the medical imaging analysis domain, where data scarcity limits possible performance gains for deep learning models. Some advancements have been made in boosting the transfer learning performance gain by merging models starting from the same initialization. However, in the medical imaging analysis domain, there is an opportunity in merging models starting from different initialisations, thus combining the features learnt from different tasks. In this work, we propose MedMerge, a method whereby the weights of different models can be merged, and their features can be effectively utilized to boost performance on a new task. With MedMerge, we learn kernel-level weights that can later be used to merge the models into a single model, even when starting from different initializations. Testing on various medical imaging analysis tasks, we show that our merged model can achieve significant performance gains, with up to 3% improvement on the F1 score. The code implementation of this work will be available at www.github.com/BioMedIA-MBZUAI/MedMerge.
XReal: Realistic Anatomy and Pathology-Aware X-ray Generation via Controllable Diffusion Model
Mar 14, 2024



Abstract:Large-scale generative models have demonstrated impressive capacity in producing visually compelling images, with increasing applications in medical imaging. However, they continue to grapple with the challenge of image hallucination and the generation of anatomically inaccurate outputs. These limitations are mainly due to the sole reliance on textual inputs and lack of spatial control over the generated images, hindering the potential usefulness of such models in real-life settings. We present XReal, a novel controllable diffusion model for generating realistic chest X-ray images through precise anatomy and pathology location control. Our lightweight method can seamlessly integrate spatial control in a pre-trained text-to-image diffusion model without fine-tuning, retaining its existing knowledge while enhancing its generation capabilities. XReal outperforms state-of-the-art x-ray diffusion models in quantitative and qualitative metrics while showing 13% and 10% anatomy and pathology realism gain, respectively, based on the expert radiologist evaluation. Our model holds promise for advancing generative models in medical imaging, offering greater precision and adaptability while inviting further exploration in this evolving field. A large synthetically generated data with annotations and code is publicly available at https://github.com/BioMedIA-MBZUAI/XReal.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge