Sarim Hashmi
Robust and Calibrated Detection of Authentic Multimedia Content
Dec 17, 2025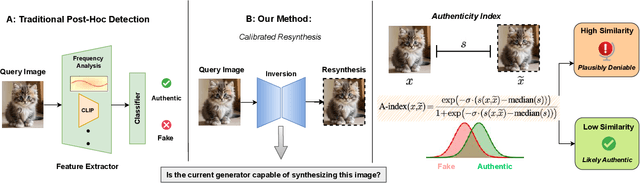
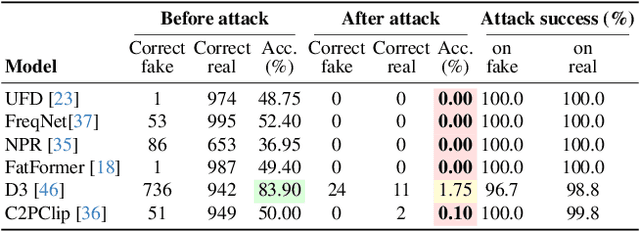
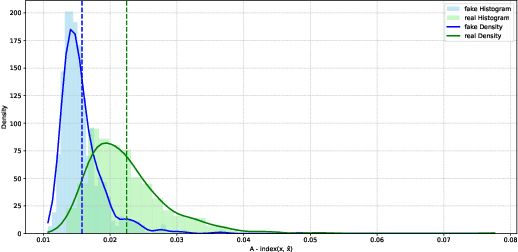
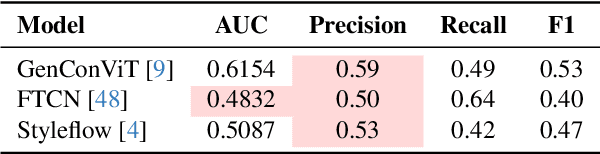
Abstract:Generative models can synthesize highly realistic content, so-called deepfakes, that are already being misused at scale to undermine digital media authenticity. Current deepfake detection methods are unreliable for two reasons: (i) distinguishing inauthentic content post-hoc is often impossible (e.g., with memorized samples), leading to an unbounded false positive rate (FPR); and (ii) detection lacks robustness, as adversaries can adapt to known detectors with near-perfect accuracy using minimal computational resources. To address these limitations, we propose a resynthesis framework to determine if a sample is authentic or if its authenticity can be plausibly denied. We make two key contributions focusing on the high-precision, low-recall setting against efficient (i.e., compute-restricted) adversaries. First, we demonstrate that our calibrated resynthesis method is the most reliable approach for verifying authentic samples while maintaining controllable, low FPRs. Second, we show that our method achieves adversarial robustness against efficient adversaries, whereas prior methods are easily evaded under identical compute budgets. Our approach supports multiple modalities and leverages state-of-the-art inversion techniques.
Guaranteed Guess: A Language Modeling Approach for CISC-to-RISC Transpilation with Testing Guarantees
Jun 17, 2025Abstract:The hardware ecosystem is rapidly evolving, with increasing interest in translating low-level programs across different instruction set architectures (ISAs) in a quick, flexible, and correct way to enhance the portability and longevity of existing code. A particularly challenging class of this transpilation problem is translating between complex- (CISC) and reduced- (RISC) hardware architectures, due to fundamental differences in instruction complexity, memory models, and execution paradigms. In this work, we introduce GG (Guaranteed Guess), an ISA-centric transpilation pipeline that combines the translation power of pre-trained large language models (LLMs) with the rigor of established software testing constructs. Our method generates candidate translations using an LLM from one ISA to another, and embeds such translations within a software-testing framework to build quantifiable confidence in the translation. We evaluate our GG approach over two diverse datasets, enforce high code coverage (>98%) across unit tests, and achieve functional/semantic correctness of 99% on HumanEval programs and 49% on BringupBench programs, respectively. Further, we compare our approach to the state-of-the-art Rosetta 2 framework on Apple Silicon, showcasing 1.73x faster runtime performance, 1.47x better energy efficiency, and 2.41x better memory usage for our transpiled code, demonstrating the effectiveness of GG for real-world CISC-to-RISC translation tasks. We will open-source our codes, data, models, and benchmarks to establish a common foundation for ISA-level code translation research.
CASS: Nvidia to AMD Transpilation with Data, Models, and Benchmark
May 22, 2025Abstract:We introduce \texttt{CASS}, the first large-scale dataset and model suite for cross-architecture GPU code transpilation, targeting both source-level (CUDA~$\leftrightarrow$~HIP) and assembly-level (Nvidia SASS~$\leftrightarrow$~AMD RDNA3) translation. The dataset comprises 70k verified code pairs across host and device, addressing a critical gap in low-level GPU code portability. Leveraging this resource, we train the \texttt{CASS} family of domain-specific language models, achieving 95\% source translation accuracy and 37.5\% assembly translation accuracy, substantially outperforming commercial baselines such as GPT-4o, Claude, and Hipify. Our generated code matches native performance in over 85\% of test cases, preserving runtime and memory behavior. To support rigorous evaluation, we introduce \texttt{CASS-Bench}, a curated benchmark spanning 16 GPU domains with ground-truth execution. All data, models, and evaluation tools are released as open source to foster progress in GPU compiler tooling, binary compatibility, and LLM-guided hardware translation. Dataset and benchmark are on \href{https://huggingface.co/datasets/MBZUAI/cass}{\textcolor{blue}{HuggingFace}}, with code at \href{https://github.com/GustavoStahl/CASS}{\textcolor{blue}{GitHub}}.
SALT: Singular Value Adaptation with Low-Rank Transformation
Mar 20, 2025Abstract:The complex nature of medical image segmentation calls for models that are specifically designed to capture detailed, domain-specific features. Large foundation models offer considerable flexibility, yet the cost of fine-tuning these models remains a significant barrier. Parameter-Efficient Fine-Tuning (PEFT) methods, such as Low-Rank Adaptation (LoRA), efficiently update model weights with low-rank matrices but may suffer from underfitting when the chosen rank is insufficient to capture domain-specific nuances. Conversely, full-rank Singular Value Decomposition (SVD) based methods provide comprehensive updates by modifying all singular values, yet they often lack flexibility and exhibit variable performance across datasets. We propose SALT (Singular Value Adaptation with Low-Rank Transformation), a method that selectively adapts the most influential singular values using trainable scale and shift parameters while complementing this with a low-rank update for the remaining subspace. This hybrid approach harnesses the advantages of both LoRA and SVD, enabling effective adaptation without relying on increasing model size or depth. Evaluated on 5 challenging medical datasets, ranging from as few as 20 samples to 1000, SALT outperforms state-of-the-art PEFT (LoRA and SVD) by 2% to 5% in Dice with only 3.9% trainable parameters, demonstrating robust adaptation even in low-resource settings. The code for SALT is available at: https://github.com/BioMedIA-MBZUAI/SALT
Language and Planning in Robotic Navigation: A Multilingual Evaluation of State-of-the-Art Models
Jan 07, 2025



Abstract:Large Language Models (LLMs) such as GPT-4, trained on huge amount of datasets spanning multiple domains, exhibit significant reasoning, understanding, and planning capabilities across various tasks. This study presents the first-ever work in Arabic language integration within the Vision-and-Language Navigation (VLN) domain in robotics, an area that has been notably underexplored in existing research. We perform a comprehensive evaluation of state-of-the-art multi-lingual Small Language Models (SLMs), including GPT-4o mini, Llama 3 8B, and Phi-3 medium 14B, alongside the Arabic-centric LLM, Jais. Our approach utilizes the NavGPT framework, a pure LLM-based instruction-following navigation agent, to assess the impact of language on navigation reasoning through zero-shot sequential action prediction using the R2R dataset. Through comprehensive experiments, we demonstrate that our framework is capable of high-level planning for navigation tasks when provided with instructions in both English and Arabic. However, certain models struggled with reasoning and planning in the Arabic language due to inherent limitations in their capabilities, sub-optimal performance, and parsing issues. These findings highlight the importance of enhancing planning and reasoning capabilities in language models for effective navigation, emphasizing this as a key area for further development while also unlocking the potential of Arabic-language models for impactful real-world applications.
Optimizing Brain Tumor Segmentation with MedNeXt: BraTS 2024 SSA and Pediatrics
Nov 26, 2024



Abstract:Identifying key pathological features in brain MRIs is crucial for the long-term survival of glioma patients. However, manual segmentation is time-consuming, requiring expert intervention and is susceptible to human error. Therefore, significant research has been devoted to developing machine learning methods that can accurately segment tumors in 3D multimodal brain MRI scans. Despite their progress, state-of-the-art models are often limited by the data they are trained on, raising concerns about their reliability when applied to diverse populations that may introduce distribution shifts. Such shifts can stem from lower quality MRI technology (e.g., in sub-Saharan Africa) or variations in patient demographics (e.g., children). The BraTS-2024 challenge provides a platform to address these issues. This study presents our methodology for segmenting tumors in the BraTS-2024 SSA and Pediatric Tumors tasks using MedNeXt, comprehensive model ensembling, and thorough postprocessing. Our approach demonstrated strong performance on the unseen validation set, achieving an average Dice Similarity Coefficient (DSC) of 0.896 on the BraTS-2024 SSA dataset and an average DSC of 0.830 on the BraTS Pediatric Tumor dataset. Additionally, our method achieved an average Hausdorff Distance (HD95) of 14.682 on the BraTS-2024 SSA dataset and an average HD95 of 37.508 on the BraTS Pediatric dataset. Our GitHub repository can be accessed here: Project Repository : https://github.com/python-arch/BioMbz-Optimizing-Brain-Tumor-Segmentation-with-MedNeXt-BraTS-2024-SSA-and-Pediatrics
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge