Adrien Depeursinge
for the Image Biomarker Standardisation Initiative
Benchmarking and Explaining Deep Learning Cortical Lesion MRI Segmentation in Multiple Sclerosis
Jul 16, 2025Abstract:Cortical lesions (CLs) have emerged as valuable biomarkers in multiple sclerosis (MS), offering high diagnostic specificity and prognostic relevance. However, their routine clinical integration remains limited due to subtle magnetic resonance imaging (MRI) appearance, challenges in expert annotation, and a lack of standardized automated methods. We propose a comprehensive multi-centric benchmark of CL detection and segmentation in MRI. A total of 656 MRI scans, including clinical trial and research data from four institutions, were acquired at 3T and 7T using MP2RAGE and MPRAGE sequences with expert-consensus annotations. We rely on the self-configuring nnU-Net framework, designed for medical imaging segmentation, and propose adaptations tailored to the improved CL detection. We evaluated model generalization through out-of-distribution testing, demonstrating strong lesion detection capabilities with an F1-score of 0.64 and 0.5 in and out of the domain, respectively. We also analyze internal model features and model errors for a better understanding of AI decision-making. Our study examines how data variability, lesion ambiguity, and protocol differences impact model performance, offering future recommendations to address these barriers to clinical adoption. To reinforce the reproducibility, the implementation and models will be publicly accessible and ready to use at https://github.com/Medical-Image-Analysis-Laboratory/ and https://doi.org/10.5281/zenodo.15911797.
A Multi-Centric Anthropomorphic 3D CT Phantom-Based Benchmark Dataset for Harmonization
Jul 02, 2025Abstract:Artificial intelligence (AI) has introduced numerous opportunities for human assistance and task automation in medicine. However, it suffers from poor generalization in the presence of shifts in the data distribution. In the context of AI-based computed tomography (CT) analysis, significant data distribution shifts can be caused by changes in scanner manufacturer, reconstruction technique or dose. AI harmonization techniques can address this problem by reducing distribution shifts caused by various acquisition settings. This paper presents an open-source benchmark dataset containing CT scans of an anthropomorphic phantom acquired with various scanners and settings, which purpose is to foster the development of AI harmonization techniques. Using a phantom allows fixing variations attributed to inter- and intra-patient variations. The dataset includes 1378 image series acquired with 13 scanners from 4 manufacturers across 8 institutions using a harmonized protocol as well as several acquisition doses. Additionally, we present a methodology, baseline results and open-source code to assess image- and feature-level stability and liver tissue classification, promoting the development of AI harmonization strategies.
Explainability of AI Uncertainty: Application to Multiple Sclerosis Lesion Segmentation on MRI
Apr 07, 2025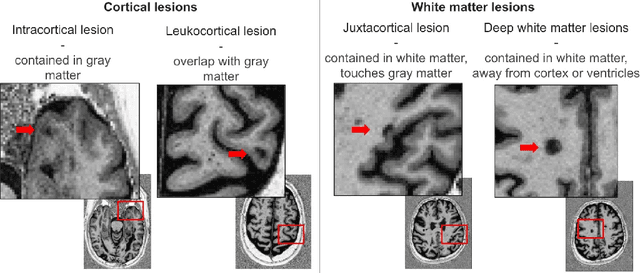



Abstract:Trustworthy artificial intelligence (AI) is essential in healthcare, particularly for high-stakes tasks like medical image segmentation. Explainable AI and uncertainty quantification significantly enhance AI reliability by addressing key attributes such as robustness, usability, and explainability. Despite extensive technical advances in uncertainty quantification for medical imaging, understanding the clinical informativeness and interpretability of uncertainty remains limited. This study introduces a novel framework to explain the potential sources of predictive uncertainty, specifically in cortical lesion segmentation in multiple sclerosis using deep ensembles. The proposed analysis shifts the focus from the uncertainty-error relationship towards relevant medical and engineering factors. Our findings reveal that instance-wise uncertainty is strongly related to lesion size, shape, and cortical involvement. Expert rater feedback confirms that similar factors impede annotator confidence. Evaluations conducted on two datasets (206 patients, almost 2000 lesions) under both in-domain and distribution-shift conditions highlight the utility of the framework in different scenarios.
Interpretability of Uncertainty: Exploring Cortical Lesion Segmentation in Multiple Sclerosis
Jul 08, 2024


Abstract:Uncertainty quantification (UQ) has become critical for evaluating the reliability of artificial intelligence systems, especially in medical image segmentation. This study addresses the interpretability of instance-wise uncertainty values in deep learning models for focal lesion segmentation in magnetic resonance imaging, specifically cortical lesion (CL) segmentation in multiple sclerosis. CL segmentation presents several challenges, including the complexity of manual segmentation, high variability in annotation, data scarcity, and class imbalance, all of which contribute to aleatoric and epistemic uncertainty. We explore how UQ can be used not only to assess prediction reliability but also to provide insights into model behavior, detect biases, and verify the accuracy of UQ methods. Our research demonstrates the potential of instance-wise uncertainty values to offer post hoc global model explanations, serving as a sanity check for the model. The implementation is available at https://github.com/NataliiaMolch/interpret-lesion-unc.
Instance-level quantitative saliency in multiple sclerosis lesion segmentation
Jun 13, 2024
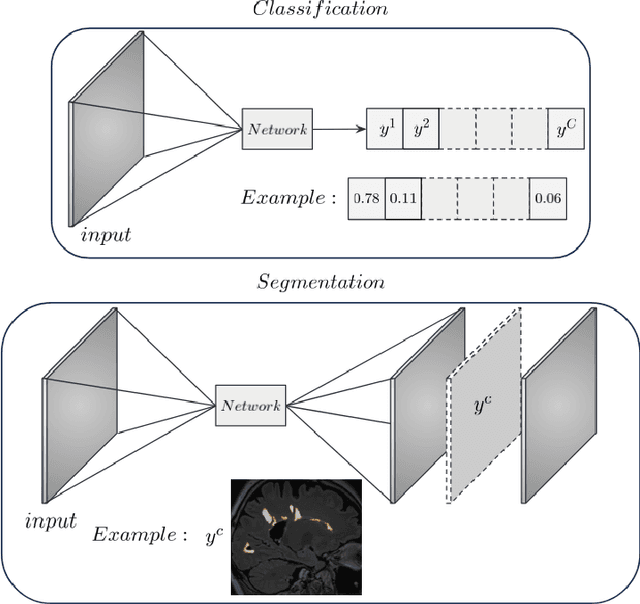


Abstract:In recent years, explainable methods for artificial intelligence (XAI) have tried to reveal and describe models' decision mechanisms in the case of classification tasks. However, XAI for semantic segmentation and in particular for single instances has been little studied to date. Understanding the process underlying automatic segmentation of single instances is crucial to reveal what information was used to detect and segment a given object of interest. In this study, we proposed two instance-level explanation maps for semantic segmentation based on SmoothGrad and Grad-CAM++ methods. Then, we investigated their relevance for the detection and segmentation of white matter lesions (WML), a magnetic resonance imaging (MRI) biomarker in multiple sclerosis (MS). 687 patients diagnosed with MS for a total of 4043 FLAIR and MPRAGE MRI scans were collected at the University Hospital of Basel, Switzerland. Data were randomly split into training, validation and test sets to train a 3D U-Net for MS lesion segmentation. We observed 3050 true positive (TP), 1818 false positive (FP), and 789 false negative (FN) cases. We generated instance-level explanation maps for semantic segmentation, by developing two XAI methods based on SmoothGrad and Grad-CAM++. We investigated: 1) the distribution of gradients in saliency maps with respect to both input MRI sequences; 2) the model's response in the case of synthetic lesions; 3) the amount of perilesional tissue needed by the model to segment a lesion. Saliency maps (based on SmoothGrad) in FLAIR showed positive values inside a lesion and negative in its neighborhood. Peak values of saliency maps generated for these four groups of volumes presented distributions that differ significantly from one another, suggesting a quantitative nature of the proposed saliency. Contextual information of 7mm around the lesion border was required for their segmentation.
EDUE: Expert Disagreement-Guided One-Pass Uncertainty Estimation for Medical Image Segmentation
Mar 25, 2024
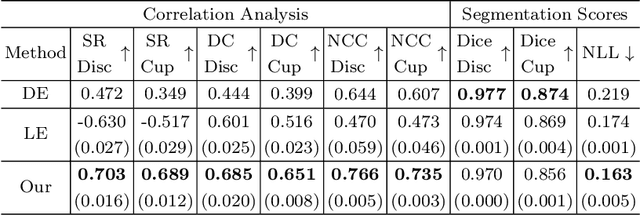
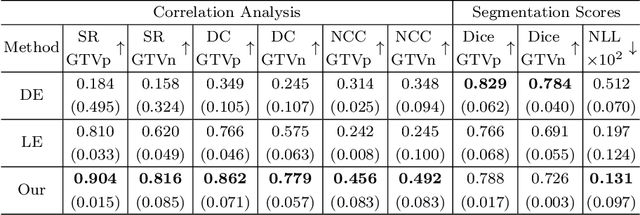

Abstract:Deploying deep learning (DL) models in medical applications relies on predictive performance and other critical factors, such as conveying trustworthy predictive uncertainty. Uncertainty estimation (UE) methods provide potential solutions for evaluating prediction reliability and improving the model confidence calibration. Despite increasing interest in UE, challenges persist, such as the need for explicit methods to capture aleatoric uncertainty and align uncertainty estimates with real-life disagreements among domain experts. This paper proposes an Expert Disagreement-Guided Uncertainty Estimation (EDUE) for medical image segmentation. By leveraging variability in ground-truth annotations from multiple raters, we guide the model during training and incorporate random sampling-based strategies to enhance calibration confidence. Our method achieves 55% and 23% improvement in correlation on average with expert disagreements at the image and pixel levels, respectively, better calibration, and competitive segmentation performance compared to the state-of-the-art deep ensembles, requiring only a single forward pass.
Structural-Based Uncertainty in Deep Learning Across Anatomical Scales: Analysis in White Matter Lesion Segmentation
Nov 15, 2023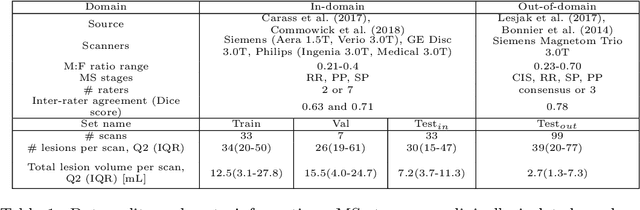


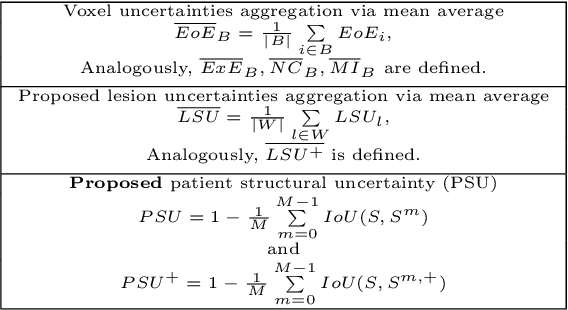
Abstract:This paper explores uncertainty quantification (UQ) as an indicator of the trustworthiness of automated deep-learning (DL) tools in the context of white matter lesion (WML) segmentation from magnetic resonance imaging (MRI) scans of multiple sclerosis (MS) patients. Our study focuses on two principal aspects of uncertainty in structured output segmentation tasks. Firstly, we postulate that a good uncertainty measure should indicate predictions likely to be incorrect with high uncertainty values. Second, we investigate the merit of quantifying uncertainty at different anatomical scales (voxel, lesion, or patient). We hypothesize that uncertainty at each scale is related to specific types of errors. Our study aims to confirm this relationship by conducting separate analyses for in-domain and out-of-domain settings. Our primary methodological contributions are (i) the development of novel measures for quantifying uncertainty at lesion and patient scales, derived from structural prediction discrepancies, and (ii) the extension of an error retention curve analysis framework to facilitate the evaluation of UQ performance at both lesion and patient scales. The results from a multi-centric MRI dataset of 172 patients demonstrate that our proposed measures more effectively capture model errors at the lesion and patient scales compared to measures that average voxel-scale uncertainty values. We provide the UQ protocols code at https://github.com/Medical-Image-Analysis-Laboratory/MS_WML_uncs.
MedShapeNet -- A Large-Scale Dataset of 3D Medical Shapes for Computer Vision
Sep 12, 2023



Abstract:We present MedShapeNet, a large collection of anatomical shapes (e.g., bones, organs, vessels) and 3D surgical instrument models. Prior to the deep learning era, the broad application of statistical shape models (SSMs) in medical image analysis is evidence that shapes have been commonly used to describe medical data. Nowadays, however, state-of-the-art (SOTA) deep learning algorithms in medical imaging are predominantly voxel-based. In computer vision, on the contrary, shapes (including, voxel occupancy grids, meshes, point clouds and implicit surface models) are preferred data representations in 3D, as seen from the numerous shape-related publications in premier vision conferences, such as the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), as well as the increasing popularity of ShapeNet (about 51,300 models) and Princeton ModelNet (127,915 models) in computer vision research. MedShapeNet is created as an alternative to these commonly used shape benchmarks to facilitate the translation of data-driven vision algorithms to medical applications, and it extends the opportunities to adapt SOTA vision algorithms to solve critical medical problems. Besides, the majority of the medical shapes in MedShapeNet are modeled directly on the imaging data of real patients, and therefore it complements well existing shape benchmarks comprising of computer-aided design (CAD) models. MedShapeNet currently includes more than 100,000 medical shapes, and provides annotations in the form of paired data. It is therefore also a freely available repository of 3D models for extended reality (virtual reality - VR, augmented reality - AR, mixed reality - MR) and medical 3D printing. This white paper describes in detail the motivations behind MedShapeNet, the shape acquisition procedures, the use cases, as well as the usage of the online shape search portal: https://medshapenet.ikim.nrw/
Why is the winner the best?
Mar 30, 2023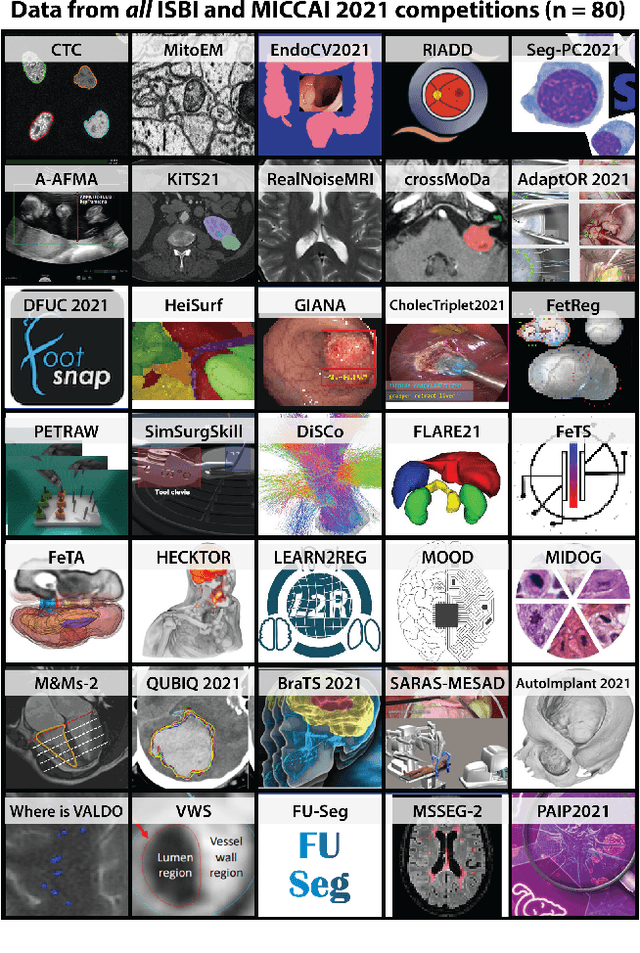
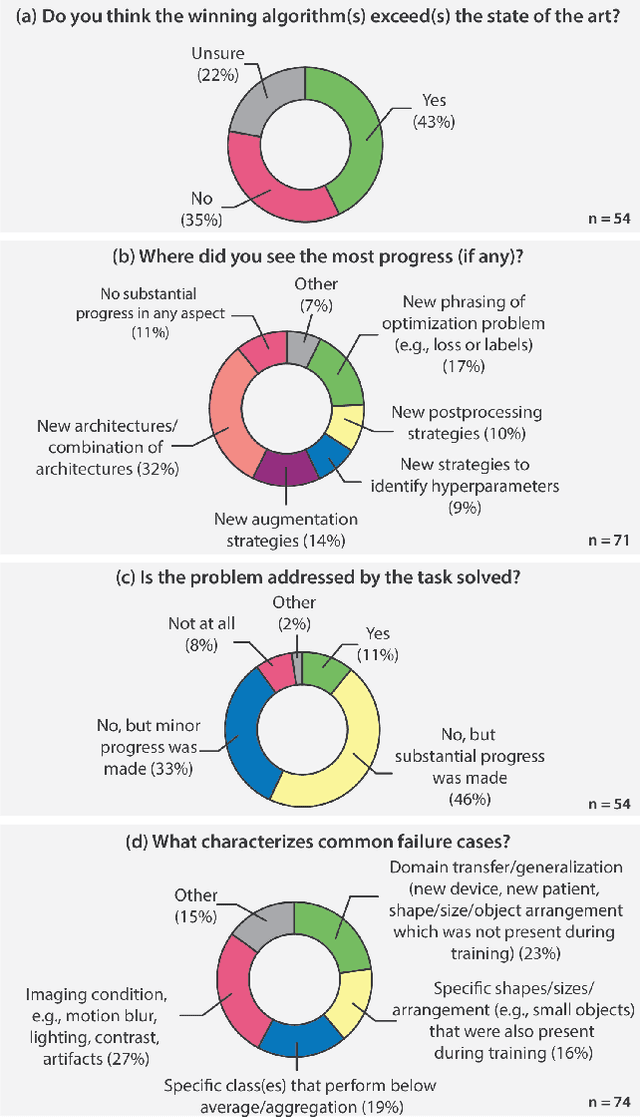
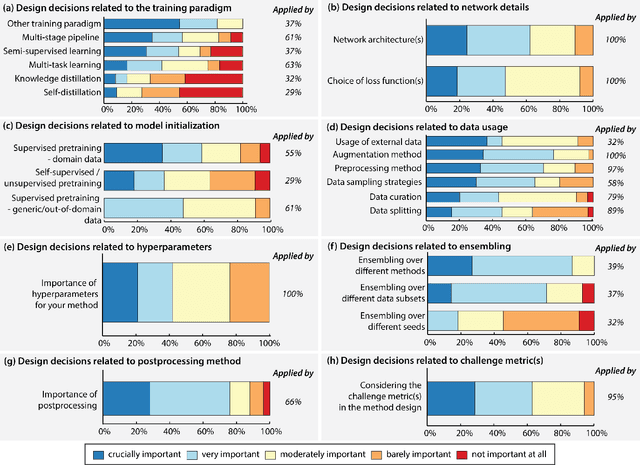
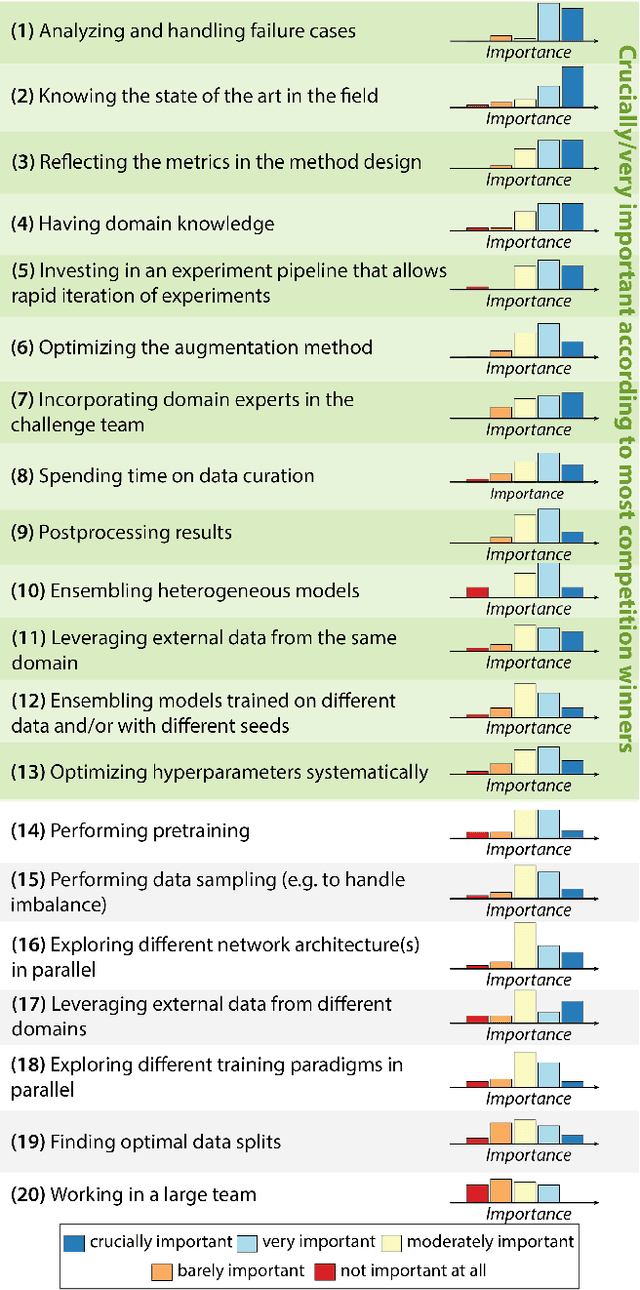
Abstract:International benchmarking competitions have become fundamental for the comparative performance assessment of image analysis methods. However, little attention has been given to investigating what can be learnt from these competitions. Do they really generate scientific progress? What are common and successful participation strategies? What makes a solution superior to a competing method? To address this gap in the literature, we performed a multi-center study with all 80 competitions that were conducted in the scope of IEEE ISBI 2021 and MICCAI 2021. Statistical analyses performed based on comprehensive descriptions of the submitted algorithms linked to their rank as well as the underlying participation strategies revealed common characteristics of winning solutions. These typically include the use of multi-task learning (63%) and/or multi-stage pipelines (61%), and a focus on augmentation (100%), image preprocessing (97%), data curation (79%), and postprocessing (66%). The "typical" lead of a winning team is a computer scientist with a doctoral degree, five years of experience in biomedical image analysis, and four years of experience in deep learning. Two core general development strategies stood out for highly-ranked teams: the reflection of the metrics in the method design and the focus on analyzing and handling failure cases. According to the organizers, 43% of the winning algorithms exceeded the state of the art but only 11% completely solved the respective domain problem. The insights of our study could help researchers (1) improve algorithm development strategies when approaching new problems, and (2) focus on open research questions revealed by this work.
Biomedical image analysis competitions: The state of current participation practice
Dec 16, 2022Abstract:The number of international benchmarking competitions is steadily increasing in various fields of machine learning (ML) research and practice. So far, however, little is known about the common practice as well as bottlenecks faced by the community in tackling the research questions posed. To shed light on the status quo of algorithm development in the specific field of biomedical imaging analysis, we designed an international survey that was issued to all participants of challenges conducted in conjunction with the IEEE ISBI 2021 and MICCAI 2021 conferences (80 competitions in total). The survey covered participants' expertise and working environments, their chosen strategies, as well as algorithm characteristics. A median of 72% challenge participants took part in the survey. According to our results, knowledge exchange was the primary incentive (70%) for participation, while the reception of prize money played only a minor role (16%). While a median of 80 working hours was spent on method development, a large portion of participants stated that they did not have enough time for method development (32%). 25% perceived the infrastructure to be a bottleneck. Overall, 94% of all solutions were deep learning-based. Of these, 84% were based on standard architectures. 43% of the respondents reported that the data samples (e.g., images) were too large to be processed at once. This was most commonly addressed by patch-based training (69%), downsampling (37%), and solving 3D analysis tasks as a series of 2D tasks. K-fold cross-validation on the training set was performed by only 37% of the participants and only 50% of the participants performed ensembling based on multiple identical models (61%) or heterogeneous models (39%). 48% of the respondents applied postprocessing steps.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge