Roger Schaer
for the Image Biomarker Standardisation Initiative
A Multi-Centric Anthropomorphic 3D CT Phantom-Based Benchmark Dataset for Harmonization
Jul 02, 2025Abstract:Artificial intelligence (AI) has introduced numerous opportunities for human assistance and task automation in medicine. However, it suffers from poor generalization in the presence of shifts in the data distribution. In the context of AI-based computed tomography (CT) analysis, significant data distribution shifts can be caused by changes in scanner manufacturer, reconstruction technique or dose. AI harmonization techniques can address this problem by reducing distribution shifts caused by various acquisition settings. This paper presents an open-source benchmark dataset containing CT scans of an anthropomorphic phantom acquired with various scanners and settings, which purpose is to foster the development of AI harmonization techniques. Using a phantom allows fixing variations attributed to inter- and intra-patient variations. The dataset includes 1378 image series acquired with 13 scanners from 4 manufacturers across 8 institutions using a harmonized protocol as well as several acquisition doses. Additionally, we present a methodology, baseline results and open-source code to assess image- and feature-level stability and liver tissue classification, promoting the development of AI harmonization strategies.
Instance-level quantitative saliency in multiple sclerosis lesion segmentation
Jun 13, 2024
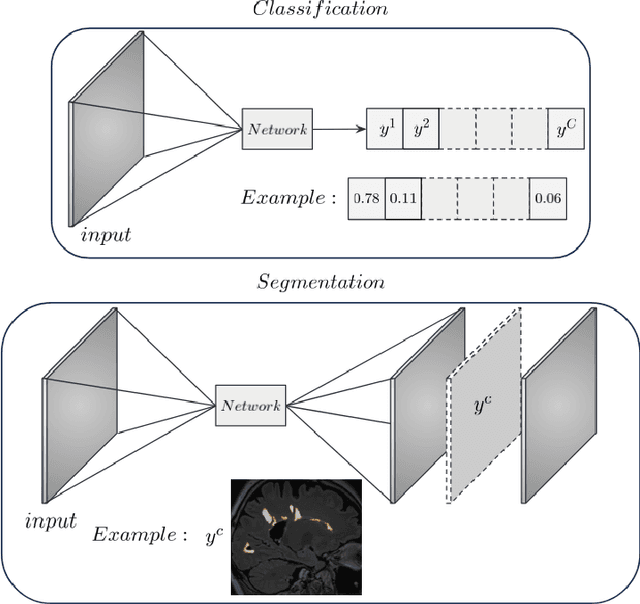


Abstract:In recent years, explainable methods for artificial intelligence (XAI) have tried to reveal and describe models' decision mechanisms in the case of classification tasks. However, XAI for semantic segmentation and in particular for single instances has been little studied to date. Understanding the process underlying automatic segmentation of single instances is crucial to reveal what information was used to detect and segment a given object of interest. In this study, we proposed two instance-level explanation maps for semantic segmentation based on SmoothGrad and Grad-CAM++ methods. Then, we investigated their relevance for the detection and segmentation of white matter lesions (WML), a magnetic resonance imaging (MRI) biomarker in multiple sclerosis (MS). 687 patients diagnosed with MS for a total of 4043 FLAIR and MPRAGE MRI scans were collected at the University Hospital of Basel, Switzerland. Data were randomly split into training, validation and test sets to train a 3D U-Net for MS lesion segmentation. We observed 3050 true positive (TP), 1818 false positive (FP), and 789 false negative (FN) cases. We generated instance-level explanation maps for semantic segmentation, by developing two XAI methods based on SmoothGrad and Grad-CAM++. We investigated: 1) the distribution of gradients in saliency maps with respect to both input MRI sequences; 2) the model's response in the case of synthetic lesions; 3) the amount of perilesional tissue needed by the model to segment a lesion. Saliency maps (based on SmoothGrad) in FLAIR showed positive values inside a lesion and negative in its neighborhood. Peak values of saliency maps generated for these four groups of volumes presented distributions that differ significantly from one another, suggesting a quantitative nature of the proposed saliency. Contextual information of 7mm around the lesion border was required for their segmentation.
Standardised convolutional filtering for radiomics
Jun 09, 2020



Abstract:The Image Biomarker Standardisation Initiative (IBSI) aims to improve reproducibility of radiomics studies by standardising the computational process of extracting image biomarkers (features) from images. We have previously established reference values for 169 commonly used features, created a standard radiomics image processing scheme, and developed reporting guidelines for radiomic studies. However, several aspects are not standardised. Here we present a preliminary version of a reference manual on the use of convolutional image filters in radiomics. Filters, such as wavelets or Laplacian of Gaussian filters, play an important part in emphasising specific image characteristics such as edges and blobs. Features derived from filter response maps have been found to be poorly reproducible. This reference manual forms the basis of ongoing work on standardising convolutional filters in radiomics, and will be updated as this work progresses.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge