Nataliia Molchanova
From 100,000+ images to winning the first brain MRI foundation model challenges: Sharing lessons and models
Jan 19, 2026Abstract:Developing Foundation Models for medical image analysis is essential to overcome the unique challenges of radiological tasks. The first challenges of this kind for 3D brain MRI, SSL3D and FOMO25, were held at MICCAI 2025. Our solution ranked first in tracks of both contests. It relies on a U-Net CNN architecture combined with strategies leveraging anatomical priors and neuroimaging domain knowledge. Notably, our models trained 1-2 orders of magnitude faster and were 10 times smaller than competing transformer-based approaches. Models are available here: https://github.com/jbanusco/BrainFM4Challenges.
Causal Attribution of Model Performance Gaps in Medical Imaging Under Distribution Shifts
Dec 09, 2025Abstract:Deep learning models for medical image segmentation suffer significant performance drops due to distribution shifts, but the causal mechanisms behind these drops remain poorly understood. We extend causal attribution frameworks to high-dimensional segmentation tasks, quantifying how acquisition protocols and annotation variability independently contribute to performance degradation. We model the data-generating process through a causal graph and employ Shapley values to fairly attribute performance changes to individual mechanisms. Our framework addresses unique challenges in medical imaging: high-dimensional outputs, limited samples, and complex mechanism interactions. Validation on multiple sclerosis (MS) lesion segmentation across 4 centers and 7 annotators reveals context-dependent failure modes: annotation protocol shifts dominate when crossing annotators (7.4% $\pm$ 8.9% DSC attribution), while acquisition shifts dominate when crossing imaging centers (6.5% $\pm$ 9.1%). This mechanism-specific quantification enables practitioners to prioritize targeted interventions based on deployment context.
Benchmarking and Explaining Deep Learning Cortical Lesion MRI Segmentation in Multiple Sclerosis
Jul 16, 2025Abstract:Cortical lesions (CLs) have emerged as valuable biomarkers in multiple sclerosis (MS), offering high diagnostic specificity and prognostic relevance. However, their routine clinical integration remains limited due to subtle magnetic resonance imaging (MRI) appearance, challenges in expert annotation, and a lack of standardized automated methods. We propose a comprehensive multi-centric benchmark of CL detection and segmentation in MRI. A total of 656 MRI scans, including clinical trial and research data from four institutions, were acquired at 3T and 7T using MP2RAGE and MPRAGE sequences with expert-consensus annotations. We rely on the self-configuring nnU-Net framework, designed for medical imaging segmentation, and propose adaptations tailored to the improved CL detection. We evaluated model generalization through out-of-distribution testing, demonstrating strong lesion detection capabilities with an F1-score of 0.64 and 0.5 in and out of the domain, respectively. We also analyze internal model features and model errors for a better understanding of AI decision-making. Our study examines how data variability, lesion ambiguity, and protocol differences impact model performance, offering future recommendations to address these barriers to clinical adoption. To reinforce the reproducibility, the implementation and models will be publicly accessible and ready to use at https://github.com/Medical-Image-Analysis-Laboratory/ and https://doi.org/10.5281/zenodo.15911797.
Explainability of AI Uncertainty: Application to Multiple Sclerosis Lesion Segmentation on MRI
Apr 07, 2025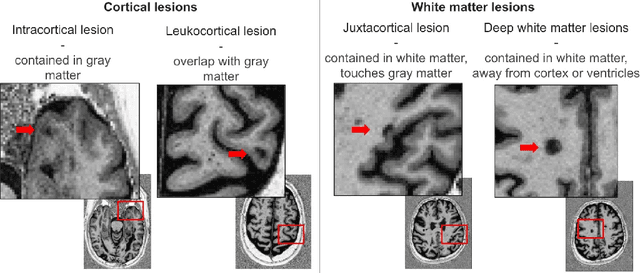



Abstract:Trustworthy artificial intelligence (AI) is essential in healthcare, particularly for high-stakes tasks like medical image segmentation. Explainable AI and uncertainty quantification significantly enhance AI reliability by addressing key attributes such as robustness, usability, and explainability. Despite extensive technical advances in uncertainty quantification for medical imaging, understanding the clinical informativeness and interpretability of uncertainty remains limited. This study introduces a novel framework to explain the potential sources of predictive uncertainty, specifically in cortical lesion segmentation in multiple sclerosis using deep ensembles. The proposed analysis shifts the focus from the uncertainty-error relationship towards relevant medical and engineering factors. Our findings reveal that instance-wise uncertainty is strongly related to lesion size, shape, and cortical involvement. Expert rater feedback confirms that similar factors impede annotator confidence. Evaluations conducted on two datasets (206 patients, almost 2000 lesions) under both in-domain and distribution-shift conditions highlight the utility of the framework in different scenarios.
Interpretability of Uncertainty: Exploring Cortical Lesion Segmentation in Multiple Sclerosis
Jul 08, 2024


Abstract:Uncertainty quantification (UQ) has become critical for evaluating the reliability of artificial intelligence systems, especially in medical image segmentation. This study addresses the interpretability of instance-wise uncertainty values in deep learning models for focal lesion segmentation in magnetic resonance imaging, specifically cortical lesion (CL) segmentation in multiple sclerosis. CL segmentation presents several challenges, including the complexity of manual segmentation, high variability in annotation, data scarcity, and class imbalance, all of which contribute to aleatoric and epistemic uncertainty. We explore how UQ can be used not only to assess prediction reliability but also to provide insights into model behavior, detect biases, and verify the accuracy of UQ methods. Our research demonstrates the potential of instance-wise uncertainty values to offer post hoc global model explanations, serving as a sanity check for the model. The implementation is available at https://github.com/NataliiaMolch/interpret-lesion-unc.
Instance-level quantitative saliency in multiple sclerosis lesion segmentation
Jun 13, 2024
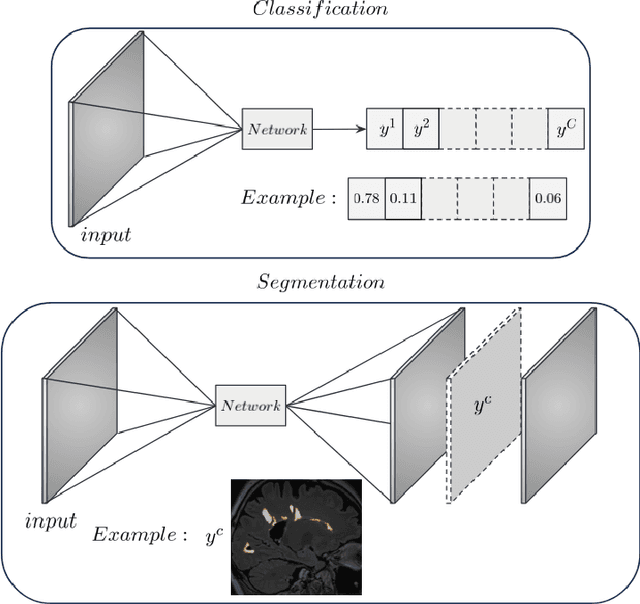


Abstract:In recent years, explainable methods for artificial intelligence (XAI) have tried to reveal and describe models' decision mechanisms in the case of classification tasks. However, XAI for semantic segmentation and in particular for single instances has been little studied to date. Understanding the process underlying automatic segmentation of single instances is crucial to reveal what information was used to detect and segment a given object of interest. In this study, we proposed two instance-level explanation maps for semantic segmentation based on SmoothGrad and Grad-CAM++ methods. Then, we investigated their relevance for the detection and segmentation of white matter lesions (WML), a magnetic resonance imaging (MRI) biomarker in multiple sclerosis (MS). 687 patients diagnosed with MS for a total of 4043 FLAIR and MPRAGE MRI scans were collected at the University Hospital of Basel, Switzerland. Data were randomly split into training, validation and test sets to train a 3D U-Net for MS lesion segmentation. We observed 3050 true positive (TP), 1818 false positive (FP), and 789 false negative (FN) cases. We generated instance-level explanation maps for semantic segmentation, by developing two XAI methods based on SmoothGrad and Grad-CAM++. We investigated: 1) the distribution of gradients in saliency maps with respect to both input MRI sequences; 2) the model's response in the case of synthetic lesions; 3) the amount of perilesional tissue needed by the model to segment a lesion. Saliency maps (based on SmoothGrad) in FLAIR showed positive values inside a lesion and negative in its neighborhood. Peak values of saliency maps generated for these four groups of volumes presented distributions that differ significantly from one another, suggesting a quantitative nature of the proposed saliency. Contextual information of 7mm around the lesion border was required for their segmentation.
Structural-Based Uncertainty in Deep Learning Across Anatomical Scales: Analysis in White Matter Lesion Segmentation
Nov 15, 2023


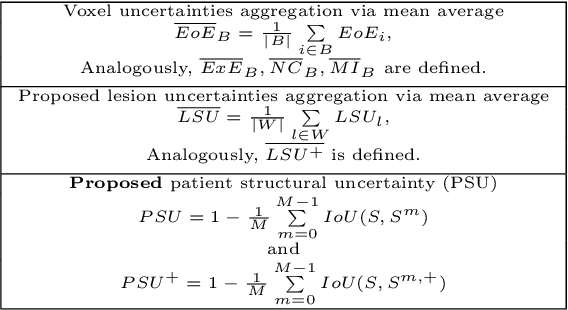
Abstract:This paper explores uncertainty quantification (UQ) as an indicator of the trustworthiness of automated deep-learning (DL) tools in the context of white matter lesion (WML) segmentation from magnetic resonance imaging (MRI) scans of multiple sclerosis (MS) patients. Our study focuses on two principal aspects of uncertainty in structured output segmentation tasks. Firstly, we postulate that a good uncertainty measure should indicate predictions likely to be incorrect with high uncertainty values. Second, we investigate the merit of quantifying uncertainty at different anatomical scales (voxel, lesion, or patient). We hypothesize that uncertainty at each scale is related to specific types of errors. Our study aims to confirm this relationship by conducting separate analyses for in-domain and out-of-domain settings. Our primary methodological contributions are (i) the development of novel measures for quantifying uncertainty at lesion and patient scales, derived from structural prediction discrepancies, and (ii) the extension of an error retention curve analysis framework to facilitate the evaluation of UQ performance at both lesion and patient scales. The results from a multi-centric MRI dataset of 172 patients demonstrate that our proposed measures more effectively capture model errors at the lesion and patient scales compared to measures that average voxel-scale uncertainty values. We provide the UQ protocols code at https://github.com/Medical-Image-Analysis-Laboratory/MS_WML_uncs.
Fast refacing of MR images with a generative neural network lowers re-identification risk and preserves volumetric consistency
May 26, 2023Abstract:With the rise of open data, identifiability of individuals based on 3D renderings obtained from routine structural magnetic resonance imaging (MRI) scans of the head has become a growing privacy concern. To protect subject privacy, several algorithms have been developed to de-identify imaging data using blurring, defacing or refacing. Completely removing facial structures provides the best re-identification protection but can significantly impact post-processing steps, like brain morphometry. As an alternative, refacing methods that replace individual facial structures with generic templates have a lower effect on the geometry and intensity distribution of original scans, and are able to provide more consistent post-processing results by the price of higher re-identification risk and computational complexity. In the current study, we propose a novel method for anonymised face generation for defaced 3D T1-weighted scans based on a 3D conditional generative adversarial network. To evaluate the performance of the proposed de-identification tool, a comparative study was conducted between several existing defacing and refacing tools, with two different segmentation algorithms (FAST and Morphobox). The aim was to evaluate (i) impact on brain morphometry reproducibility, (ii) re-identification risk, (iii) balance between (i) and (ii), and (iv) the processing time. The proposed method takes 9 seconds for face generation and is suitable for recovering consistent post-processing results after defacing.
Tackling Bias in the Dice Similarity Coefficient: Introducing nDSC for White Matter Lesion Segmentation
Feb 10, 2023Abstract:The development of automatic segmentation techniques for medical imaging tasks requires assessment metrics to fairly judge and rank such approaches on benchmarks. The Dice Similarity Coefficient (DSC) is a popular choice for comparing the agreement between the predicted segmentation against a ground-truth mask. However, the DSC metric has been shown to be biased to the occurrence rate of the positive class in the ground-truth, and hence should be considered in combination with other metrics. This work describes a detailed analysis of the recently proposed normalised Dice Similarity Coefficient (nDSC) for binary segmentation tasks as an adaptation of DSC which scales the precision at a fixed recall rate to tackle this bias. White matter lesion segmentation on magnetic resonance images of multiple sclerosis patients is selected as a case study task to empirically assess the suitability of nDSC. We validate the normalised DSC using two different models across 59 subject scans with a wide range of lesion loads. It is found that the nDSC is less biased than DSC with lesion load on standard white matter lesion segmentation benchmarks measured using standard rank correlation coefficients. An implementation of nDSC is made available at: https://github.com/NataliiaMolch/nDSC .
Novel structural-scale uncertainty measures and error retention curves: application to multiple sclerosis
Nov 11, 2022Abstract:This paper focuses on the uncertainty estimation for white matter lesions (WML) segmentation in magnetic resonance imaging (MRI). On one side, voxel-scale segmentation errors cause the erroneous delineation of the lesions; on the other side, lesion-scale detection errors lead to wrong lesion counts. Both of these factors are clinically relevant for the assessment of multiple sclerosis patients. This work aims to compare the ability of different voxel- and lesion-scale uncertainty measures to capture errors related to segmentation and lesion detection, respectively. Our main contributions are (i) proposing new measures of lesion-scale uncertainty that do not utilise voxel-scale uncertainties; (ii) extending an error retention curves analysis framework for evaluation of lesion-scale uncertainty measures. Our results obtained on the multi-center testing set of 58 patients demonstrate that the proposed lesion-scale measure achieves the best performance among the analysed measures. All code implementations are provided at https://github.com/NataliiaMolch/MS_WML_uncs
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge