Matthias Weigel
Benchmarking and Explaining Deep Learning Cortical Lesion MRI Segmentation in Multiple Sclerosis
Jul 16, 2025Abstract:Cortical lesions (CLs) have emerged as valuable biomarkers in multiple sclerosis (MS), offering high diagnostic specificity and prognostic relevance. However, their routine clinical integration remains limited due to subtle magnetic resonance imaging (MRI) appearance, challenges in expert annotation, and a lack of standardized automated methods. We propose a comprehensive multi-centric benchmark of CL detection and segmentation in MRI. A total of 656 MRI scans, including clinical trial and research data from four institutions, were acquired at 3T and 7T using MP2RAGE and MPRAGE sequences with expert-consensus annotations. We rely on the self-configuring nnU-Net framework, designed for medical imaging segmentation, and propose adaptations tailored to the improved CL detection. We evaluated model generalization through out-of-distribution testing, demonstrating strong lesion detection capabilities with an F1-score of 0.64 and 0.5 in and out of the domain, respectively. We also analyze internal model features and model errors for a better understanding of AI decision-making. Our study examines how data variability, lesion ambiguity, and protocol differences impact model performance, offering future recommendations to address these barriers to clinical adoption. To reinforce the reproducibility, the implementation and models will be publicly accessible and ready to use at https://github.com/Medical-Image-Analysis-Laboratory/ and https://doi.org/10.5281/zenodo.15911797.
Explainability of AI Uncertainty: Application to Multiple Sclerosis Lesion Segmentation on MRI
Apr 07, 2025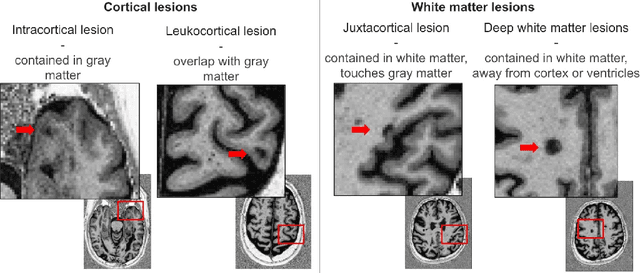



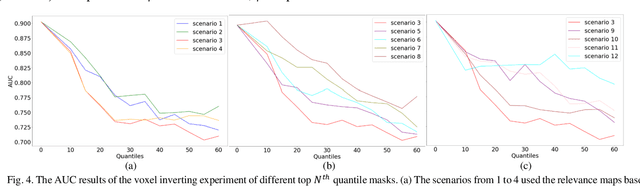
Abstract:Trustworthy artificial intelligence (AI) is essential in healthcare, particularly for high-stakes tasks like medical image segmentation. Explainable AI and uncertainty quantification significantly enhance AI reliability by addressing key attributes such as robustness, usability, and explainability. Despite extensive technical advances in uncertainty quantification for medical imaging, understanding the clinical informativeness and interpretability of uncertainty remains limited. This study introduces a novel framework to explain the potential sources of predictive uncertainty, specifically in cortical lesion segmentation in multiple sclerosis using deep ensembles. The proposed analysis shifts the focus from the uncertainty-error relationship towards relevant medical and engineering factors. Our findings reveal that instance-wise uncertainty is strongly related to lesion size, shape, and cortical involvement. Expert rater feedback confirms that similar factors impede annotator confidence. Evaluations conducted on two datasets (206 patients, almost 2000 lesions) under both in-domain and distribution-shift conditions highlight the utility of the framework in different scenarios.
Interpretability of Uncertainty: Exploring Cortical Lesion Segmentation in Multiple Sclerosis
Jul 08, 2024


Abstract:Uncertainty quantification (UQ) has become critical for evaluating the reliability of artificial intelligence systems, especially in medical image segmentation. This study addresses the interpretability of instance-wise uncertainty values in deep learning models for focal lesion segmentation in magnetic resonance imaging, specifically cortical lesion (CL) segmentation in multiple sclerosis. CL segmentation presents several challenges, including the complexity of manual segmentation, high variability in annotation, data scarcity, and class imbalance, all of which contribute to aleatoric and epistemic uncertainty. We explore how UQ can be used not only to assess prediction reliability but also to provide insights into model behavior, detect biases, and verify the accuracy of UQ methods. Our research demonstrates the potential of instance-wise uncertainty values to offer post hoc global model explanations, serving as a sanity check for the model. The implementation is available at https://github.com/NataliiaMolch/interpret-lesion-unc.
GAMER-MRIL identifies Disability-Related Brain Changes in Multiple Sclerosis
Aug 15, 2023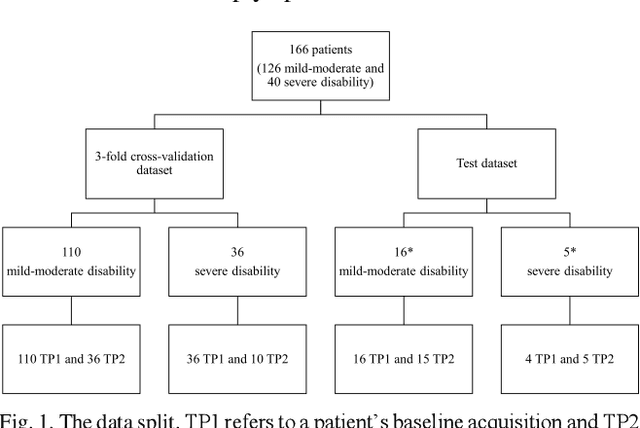
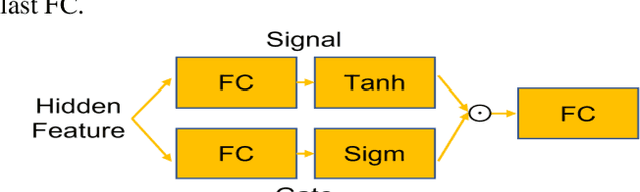
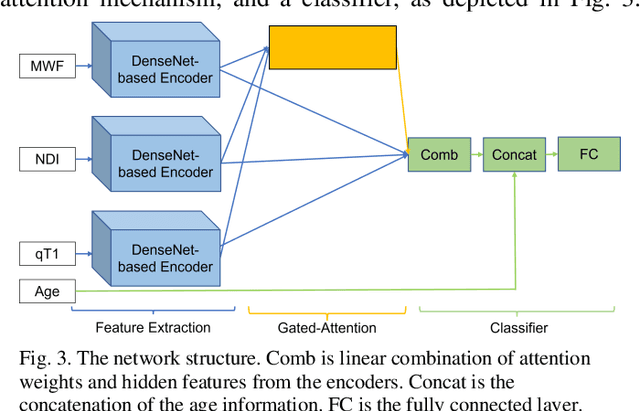
Abstract:Objective: Identifying disability-related brain changes is important for multiple sclerosis (MS) patients. Currently, there is no clear understanding about which pathological features drive disability in single MS patients. In this work, we propose a novel comprehensive approach, GAMER-MRIL, leveraging whole-brain quantitative MRI (qMRI), convolutional neural network (CNN), and an interpretability method from classifying MS patients with severe disability to investigating relevant pathological brain changes. Methods: One-hundred-sixty-six MS patients underwent 3T MRI acquisitions. qMRI informative of microstructural brain properties was reconstructed, including quantitative T1 (qT1), myelin water fraction (MWF), and neurite density index (NDI). To fully utilize the qMRI, GAMER-MRIL extended a gated-attention-based CNN (GAMER-MRI), which was developed to select patch-based qMRI important for a given task/question, to the whole-brain image. To find out disability-related brain regions, GAMER-MRIL modified a structure-aware interpretability method, Layer-wise Relevance Propagation (LRP), to incorporate qMRI. Results: The test performance was AUC=0.885. qT1 was the most sensitive measure related to disability, followed by NDI. The proposed LRP approach obtained more specifically relevant regions than other interpretability methods, including the saliency map, the integrated gradients, and the original LRP. The relevant regions included the corticospinal tract, where average qT1 and NDI significantly correlated with patients' disability scores ($\rho$=-0.37 and 0.44). Conclusion: These results demonstrated that GAMER-MRIL can classify patients with severe disability using qMRI and subsequently identify brain regions potentially important to the integrity of the mobile function. Significance: GAMER-MRIL holds promise for developing biomarkers and increasing clinicians' trust in NN.
Diffusion Models for Contrast Harmonization of Magnetic Resonance Images
Mar 14, 2023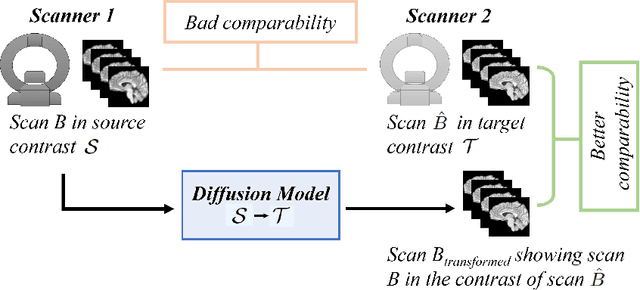
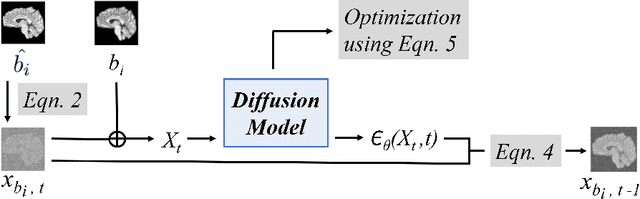
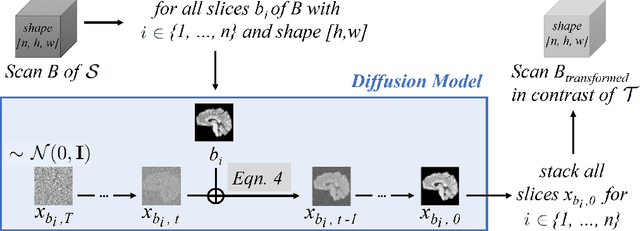

Abstract:Magnetic resonance (MR) images from multiple sources often show differences in image contrast related to acquisition settings or the used scanner type. For long-term studies, longitudinal comparability is essential but can be impaired by these contrast differences, leading to biased results when using automated evaluation tools. This study presents a diffusion model-based approach for contrast harmonization. We use a data set consisting of scans of 18 Multiple Sclerosis patients and 22 healthy controls. Each subject was scanned in two MR scanners of different magnetic field strengths (1.5 T and 3 T), resulting in a paired data set that shows scanner-inherent differences. We map images from the source contrast to the target contrast for both directions, from 3 T to 1.5 T and from 1.5 T to 3 T. As we only want to change the contrast, not the anatomical information, our method uses the original image to guide the image-to-image translation process by adding structural information. The aim is that the mapped scans display increased comparability with scans of the target contrast for downstream tasks. We evaluate this method for the task of segmentation of cerebrospinal fluid, grey matter and white matter. Our method achieves good and consistent results for both directions of the mapping.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge